Deconstructed Microfluidic Bone Marrow On-A-Chip to Study Normal and Malignant Hemopoietic Cell-Niche Interactions
- PMID: 31464364
- PMCID: PMC8011350
- DOI: 10.1002/smll.201902971
Deconstructed Microfluidic Bone Marrow On-A-Chip to Study Normal and Malignant Hemopoietic Cell-Niche Interactions
Abstract
Human hematopoietic niches are complex specialized microenvironments that maintain and regulate hematopoietic stem and progenitor cells (HSPC). Thus far, most of the studies performed investigating alterations of HSPC-niche dynamic interactions are conducted in animal models. Herein, organ microengineering with microfluidics is combined to develop a human bone marrow (BM)-on-a-chip with an integrated recirculating perfusion system that consolidates a variety of important parameters such as 3D architecture, cell-cell/cell-matrix interactions, and circulation, allowing a better mimicry of in vivo conditions. The complex BM environment is deconvoluted to 4 major distinct, but integrated, tissue-engineered 3D niche constructs housed within a single, closed, recirculating microfluidic device system, and equipped with cell tracking technology. It is shown that this technology successfully enables the identification and quantification of preferential interactions-homing and retention-of circulating normal and malignant HSPC with distinct niches.
Keywords: bone marrow niche; hematopoietic stem and progenitor cells; homing; microfluidics; tissue chip.
© 2019 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures
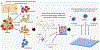





References
-
- Acar M, Kocherlakota KS, Murphy MM, Peyer JG, Oguro H, Inra CN, Jaiyeola C, Zhao Z, Luby-Phelps K, Morrison SJ, Nature 2015, 526, 126; - PMC - PubMed
- Ding L, Saunders TL, Enikolopov G, Morrison SJ, Nature 2012, 481, 457; - PMC - PubMed
- Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, Mizoguchi T, Wei Q, Lucas D, Ito K, Mar JC, Bergman A, Frenette PS, Nature 2013, 502, 637; - PMC - PubMed
- Mendelson A, Frenette PS, Nat. Med 2014, 20, 833; - PMC - PubMed
- Morrison SJ, Scadden DT, Nature 2014, 505, 327. - PMC - PubMed
-
- Crane GM, Jeffery E, Morrison SJ, Nat. Rev. Immunol 2017, 17, 573. - PubMed
-
- Horn PA, Thomasson BM, Wood BL, Andrews RG, Morris JC, Kiem HP, Blood 2003, 102, 4329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

