Ecological and evolutionary patterns in the enigmatic protist genus Percolomonas (Heterolobosea; Discoba) from diverse habitats
- PMID: 31465455
- PMCID: PMC6715209
- DOI: 10.1371/journal.pone.0216188
Ecological and evolutionary patterns in the enigmatic protist genus Percolomonas (Heterolobosea; Discoba) from diverse habitats
Abstract
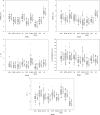
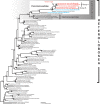
The heterotrophic flagellate Percolomonas cosmopolitus (Heterolobosea) is often observed in saline habitats worldwide, from coastal waters to saturated brines. However, only two cultures assigned to this morphospecies have been examined using molecular methods, and their 18S rRNA gene sequences are extremely different. Further the salinity tolerances of individual strains are unknown. Thus, our knowledge on the autecology and diversity in this morphospecies is deficient. Here, we report 18S rRNA gene data on seven strains similar to P. cosmopolitus from seven geographically remote locations (New Zealand, Kenya, Korea, Poland, Russia, Spain, and the USA) with sample salinities ranging from 4‰ to 280‰, and compare morphology and salinity tolerance of the nine available strains. Percolomonas cosmopolitus-like strains show few-to-no consistent morphological differences, and form six clades separated by often extremely large 18S rRNA gene divergences (up to 42.4%). Some strains grow best at salinities from 75 to 125‰ and represent halophiles. All but one of these belong to two geographically heterogeneous clusters that form a robust monophyletic group in phylogenetic trees; this likely represents an ecologically specialized subclade of halophiles. Our results suggest that P. cosmopolitus is a cluster of several cryptic species (at least), which are unlikely to be distinguished by geography. Interestingly, the 9 Percolomonas strains formed a clade in 18S rRNA gene phylogenies, unlike most previous analyses based on two sequences.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Trewick SA. Sympatric cryptic species in New Zealand Onychophora. Biol J Linnean Soc. 1998; 63:307–329.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

