Capture and Detection of Circulating Glioma Cells Using the Recombinant VAR2CSA Malaria Protein
- PMID: 31466397
- PMCID: PMC6769911
- DOI: 10.3390/cells8090998
Capture and Detection of Circulating Glioma Cells Using the Recombinant VAR2CSA Malaria Protein
Abstract
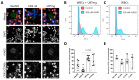
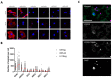
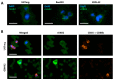
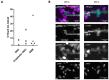
Diffuse gliomas are the most common primary malignant brain tumor. Although extracranial metastases are rarely observed, recent studies have shown the presence of circulating tumor cells (CTCs) in the blood of glioma patients, confirming that a subset of tumor cells are capable of entering the circulation. The isolation and characterization of CTCs could provide a non-invasive method for repeated analysis of the mutational and phenotypic state of the tumor during the course of disease. However, the efficient detection of glioma CTCs has proven to be challenging due to the lack of consistently expressed tumor markers and high inter- and intra-tumor heterogeneity. Thus, for this field to progress, an omnipresent but specific marker of glioma CTCs is required. In this article, we demonstrate how the recombinant malaria VAR2CSA protein (rVAR2) can be used for the capture and detection of glioma cell lines that are spiked into blood through binding to a cancer-specific oncofetal chondroitin sulfate (ofCS). When using rVAR2 pull-down from glioma cells, we identified a panel of proteoglycans, known to be essential for glioma progression. Finally, the clinical feasibility of this work is supported by the rVAR2-based isolation and detection of CTCs from glioma patient blood samples, which highlights ofCS as a potential clinical target for CTC isolation.
Keywords: biomarker; circulating tumor cells (CTCs); enrichment and detection technologies; glioma; malaria; rVAR2.
Conflict of interest statement
The technology to diagnose cancer through rVAR2 is owned by VarCT Diagnostics through a license from VAR2Pharmaceuticals. AS, MØA, TGT, MD and TMC are cofounders of VAR2Pharmaceuticals.
Figures


References
-
- Wesseling P., Kros J.M., Jeuken J.W.J.D.H. The pathological diagnosis of diffuse gliomas: Towards a smart synthesis of microscopic and molecular information in a multidisciplinary context. Diagn. Histopathol. 2011;17:486–494. doi: 10.1016/j.mpdhp.2011.08.005. - DOI
-
- Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

