Differential nanoscale organisation of LFA-1 modulates T-cell migration
- PMID: 31471459
- PMCID: PMC7614863
- DOI: 10.1242/jcs.232991
Differential nanoscale organisation of LFA-1 modulates T-cell migration
Abstract
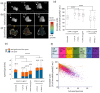
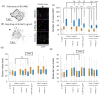
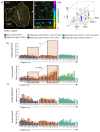
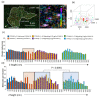
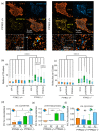
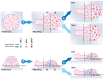
Effector T-cells rely on integrins to drive adhesion and migration to facilitate their immune function. The heterodimeric transmembrane integrin LFA-1 (αLβ2 integrin) regulates adhesion and migration of effector T-cells through linkage of the extracellular matrix with the intracellular actin treadmill machinery. Here, we quantified the velocity and direction of F-actin flow in migrating T-cells alongside single-molecule localisation of transmembrane and intracellular LFA-1. Results showed that actin retrograde flow positively correlated and immobile actin negatively correlated with T-cell velocity. Plasma membrane-localised LFA-1 forms unique nano-clustering patterns in the leading edge, compared to the mid-focal zone, of migrating T-cells. Deleting the cytosolic phosphatase PTPN22, loss-of-function mutations of which have been linked to autoimmune disease, increased T-cell velocity, and leading-edge co-clustering of pY397 FAK, pY416 Src family kinases and LFA-1. These data suggest that differential nanoclustering patterns of LFA-1 in migrating T-cells may instruct intracellular signalling. Our data presents a paradigm where T-cells modulate the nanoscale organisation of adhesion and signalling molecules to fine tune their migration speed, with implications for the regulation of immune and inflammatory responses.This article has an associated First Person interview with the first author of the paper.
Keywords: Integrins; LFA-1; SMLM; T-cell migration; dSTORM.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
References
-
- Alexandrova AY, et al. Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. [Accessed April 2, 2014];PloS one. 2008 3(9):e3234. Available at: http://www.plosone.org/article/info:doi/10.1371/journal.pone.0003234#pon... . - DOI - PMC - PubMed
-
- Begovich AB, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. [Accessed May 31, 2014];American journal of human genetics. 2004 75(2):330–7. Available at: http://www.sciencedirect.com/science/article/pii/S0002929707624161 . - PMC - PubMed
-
- Bottini N, et al. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. [Accessed June 3, 2014];Seminars in immunology. 2006 18(4):207–13. Available at: http://www.sciencedirect.com/science/article/pii/S1044532306000455 . - PubMed
-
- Brand O, Gough S, Heward J. <em>HLA</em>, <em>CTLA-4</em> and <em>PTPN22</em>: the shared genetic master-key to autoimmunity? [Accessed July 29, 2014];Expert Reviews in Molecular Medicine. 2005 7(23):1–15. Available at: http://journals.cambridge.org/abstract_S1462399405009981 . - PubMed
-
- Brown CM, et al. Probing the integrin-actin linkage using high-resolution protein velocity mapping. [Accessed April 1, 2014];Journal of cell science. 2006 119(Pt 24):5204–14. Available at: http://jcs.biologists.org/content/119/24/5204.short . - PubMed

