Proteomics, Glycomics, and Glycoproteomics of Matrisome Molecules
- PMID: 31471497
- PMCID: PMC6823855
- DOI: 10.1074/mcp.R119.001543
Proteomics, Glycomics, and Glycoproteomics of Matrisome Molecules
Abstract
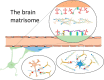
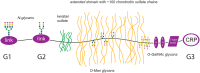
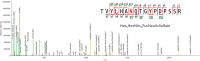
The most straightforward applications of proteomics database searching involve intracellular proteins. Although intracellular gene products number in the thousands, their well-defined post-translational modifications (PTMs) makes database searching practical. By contrast, cell surface and extracellular matrisome proteins pass through the secretory pathway where many become glycosylated, modulating their physicochemical properties, adhesive interactions, and diversifying their functions. Although matrisome proteins number only a few hundred, their high degree of complex glycosylation multiplies the number of theoretical proteoforms by orders of magnitude. Given that extracellular networks that mediate cell-cell and cell-pathogen interactions in physiology depend on glycosylation, it is important to characterize the proteomes, glycomes, and glycoproteomes of matrisome molecules that exist in a given biological context. In this review, we summarize proteomics approaches for characterizing matrisome molecules, with an emphasis on applications to brain diseases. We demonstrate the availability of methods that should greatly increase the availability of information on matrisome molecular structure associated with health and disease.
Keywords: Extracellular matrix; glycomics; glycoproteins; glycoproteomics; n-glycosylation.
© 2019 Raghunathan et al.
Figures

References
-
- Demetriou M., Nabi I. R., and Dennis J. W. (2018) Galectins as adaptors: linking glycosylation and metabolism with extracellular cues. Trends in Glycosci. Glycotechnol. 30, SE167–SE177
-
- Dennis J. W. (2017) Genetic code asymmetry supports diversity through experimentation with posttranslational modifications. Curr. Opin. Chem. Biol. 41, 1–11 - PubMed
-
- Deribe Y. L., Pawson T., and Dikic I. (2010) Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672 - PubMed
-
- Reinhardt H. C., and Yaffe M. B. (2013) Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat. Rev. Mol. Cell Biol. 14, 563–580 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

