Transcriptome profiling reveals candidate flavonol-related genes of Tetrastigma hemsleyanum under cold stress
- PMID: 31472675
- PMCID: PMC6717372
- DOI: 10.1186/s12864-019-6045-y
Transcriptome profiling reveals candidate flavonol-related genes of Tetrastigma hemsleyanum under cold stress
Abstract
Background: Tetrastigma hemsleyanum Diels et Gilg is a valuable medicinal herb, whose main bioactive constituents are flavonoids. Chilling sensitivity is the dominant environmental factor limiting growth and development of the plants. But the mechanisms of cold sensitivity in this plant are still unclear. Also, not enough information on genes involved in flavonoid biosynthesis in T. hemsleyanum is available to understand the mechanisms of its physiological and pharmaceutical effects.
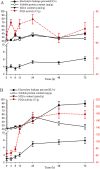
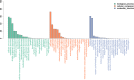
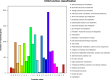
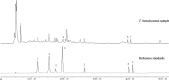
Results: The electrolyte leakage, POD activity, soluble protein, and MDA content showed a linear sustained increase under cold stress. The critical period of cold damage in T. hemsleyanum was from 12 h to 48 h. Expression profiles revealed 18,104 differentially expressed genes (DEGs) among these critical time points. Most of the cold regulated DEGs were early-response genes. A total of 114 unigenes were assigned to the flavonoid biosynthetic pathway. Fourteen genes most likely to encode flavonoid biosynthetic enzymes were identified. Flavonols of T. hemsleyanum might play a crucial role in combating cold stress. Genes encoding PAL, 4CL, CHS, ANR, FLS, and LAR were significantly up-regulated by cold stress, which could result in a significant increase in crucial flavonols (catechin, epicatechin, rutin, and quercetin) in T. hemsleyanum.
Conclusions: Overall, our results show that the expression of genes related to flavonol biosynthesis as well as flavonol content increased in T. hemsleyanum under cold stress. These findings provide valuable information regarding the transcriptome changes in response to cold stress and give a clue for identifying candidate genes as promising targets that could be used for improving cold tolerance via molecular breeding. The study also provides candidate genes involved in flavonoid biosynthesis and may be useful for clarifying the biosynthetic pathway of flavonoids in T. hemsleyanum.
Keywords: Cold stress; Flavonol; Tetrastigma hemsleyanum; Transcriptome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Transcriptome and Metabonomics Combined Analysis Revealed the Defense Mechanism Involved in Hydrogen-Rich Water-Regulated Cold Stress Response of Tetrastigma hemsleyanum.Front Plant Sci. 2022 Jun 23;13:889726. doi: 10.3389/fpls.2022.889726. eCollection 2022. Front Plant Sci. 2022. PMID: 35812920 Free PMC article.
-
Phylogenetic Analysis of R2R3-MYB Family Genes in Tetrastigma hemsleyanum Diels et Gilg and Roles of ThMYB4 and ThMYB7 in Flavonoid Biosynthesis.Biomolecules. 2023 Mar 15;13(3):531. doi: 10.3390/biom13030531. Biomolecules. 2023. PMID: 36979467 Free PMC article.
-
Flavonoid Metabolism in Tetrastigma hemsleyanum Diels et Gilg Based on Metabolome Analysis and Transcriptome Sequencing.Molecules. 2022 Dec 22;28(1):83. doi: 10.3390/molecules28010083. Molecules. 2022. PMID: 36615276 Free PMC article.
-
The research progresses and future prospects of Tetrastigma hemsleyanum Diels et Gilg: A valuable Chinese herbal medicine.J Ethnopharmacol. 2021 May 10;271:113836. doi: 10.1016/j.jep.2021.113836. Epub 2021 Jan 16. J Ethnopharmacol. 2021. PMID: 33465440 Review.
-
Insights into Metabolic Engineering of Bioactive Molecules in Tetrastigma hemsleyanum Diels & Gilg: A Traditional Medicinal Herb.Curr Genomics. 2023 Oct 27;24(2):72-83. doi: 10.2174/0113892029251472230921053135. Curr Genomics. 2023. PMID: 37994327 Free PMC article. Review.
Cited by
-
The Phytochemistry, Pharmacology, and Quality Control of Tetrastigma hemsleyanum Diels & Gilg in China: A Review.Front Pharmacol. 2020 Sep 25;11:550497. doi: 10.3389/fphar.2020.550497. eCollection 2020. Front Pharmacol. 2020. PMID: 33101019 Free PMC article. Review.
-
Transcriptome and Metabonomics Combined Analysis Revealed the Defense Mechanism Involved in Hydrogen-Rich Water-Regulated Cold Stress Response of Tetrastigma hemsleyanum.Front Plant Sci. 2022 Jun 23;13:889726. doi: 10.3389/fpls.2022.889726. eCollection 2022. Front Plant Sci. 2022. PMID: 35812920 Free PMC article.
-
The Blinin Accumulation Promoted by CbMYB32 Involved in Conyza blinii Resistance to Nocturnal Low Temperature.Int J Mol Sci. 2023 Apr 12;24(8):7143. doi: 10.3390/ijms24087143. Int J Mol Sci. 2023. PMID: 37108302 Free PMC article.
-
Transcriptome and Metabolome Integrated Analysis of Two Ecotypes of Tetrastigma hemsleyanum Reveals Candidate Genes Involved in Chlorogenic Acid Accumulation.Plants (Basel). 2021 Jun 24;10(7):1288. doi: 10.3390/plants10071288. Plants (Basel). 2021. PMID: 34202839 Free PMC article.
-
Flavonoids as key players in cold tolerance: molecular insights and applications in horticultural crops.Hortic Res. 2025 Jan 2;12(4):uhae366. doi: 10.1093/hr/uhae366. eCollection 2025 Apr. Hortic Res. 2025. PMID: 40070400 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

