Dual lysosomal-mitochondrial targeting by antihistamines to eradicate leukaemic cells
- PMID: 31473184
- PMCID: PMC6796581
- DOI: 10.1016/j.ebiom.2019.08.021
Dual lysosomal-mitochondrial targeting by antihistamines to eradicate leukaemic cells
Abstract
Background: Despite great efforts to identify druggable molecular targets for AML, there remains an unmet need for more effective therapies.
Methods: An in silico screening was performed using Connectivity Maps to identify FDA-approved drugs that may revert an early leukaemic transformation gene signature. Hit compounds were validated in AML cell lines. Cytotoxic effects were assessed both in primary AML patient samples and healthy donor blood cells. Xenotransplantation assays were undertaken to determine the effect on engraftment of hit compounds. The mechanism of action responsible for the antileukaemic effect was studied focussing on lysosomes and mitochondria.
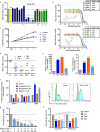
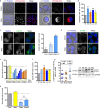
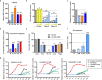
Findings: We identified a group of antihistamines (termed ANHAs) with distinct physicochemical properties associated with their cationic-amphiphilic nature, that selectively killed leukaemic cells. ANHAs behaved as antileukaemic agents against primary AML samples ex vivo, sparing healthy cells. Moreover, ANHAs severely impaired the in vivo leukaemia regeneration capacity. ANHAs' cytotoxicity relied on simultaneous mitochondrial and lysosomal disruption and induction of autophagy and apoptosis. The pharmacological effect was exerted based on their physicochemical properties that permitted the passive targeting of both organelles, without the involvement of active molecular recognition.
Interpretation: Dual targeting of lysosomes and mitochondria constitutes a new promising therapeutic approach for leukaemia treatment, supporting the further clinical development. FUND: This work was funded by the Fundación Mutua Madrileña (RMR), CaixaImpulse (RMR), the Spanish Ministry of Economy (RMR), the Josep Carreras International Leukaemia Foundation (RMR), l'Obra Social "La Caixa" (RMR), and Generalitat de Catalunya (IJC).
Keywords: Antihistamines; Ebastine; Leukaemia; Lysosomes; Mitochondria.
Copyright © 2019 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Dr. Muñoz Risueño reports grants from Ramón y Cajal program, grants from Fundación Mutua Madrileña, grants from CaixaImpulse, grants from MINECO, grants from Josep Carreras International Leukaemia Research Foundation, grants from Obra Social La Caixa-Fundació Bancària La Caixa, grants from CERCA Programme, during the conduct of the study; and RMR is a shareholder of Leukos Biotech.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

