The protein arginine methyltransferase PRMT5 confers therapeutic resistance to mTOR inhibition in glioblastoma
- PMID: 31473880
- PMCID: PMC6776692
- DOI: 10.1007/s11060-019-03274-0
The protein arginine methyltransferase PRMT5 confers therapeutic resistance to mTOR inhibition in glioblastoma
Abstract
Introduction: Clinical trials directed at mechanistic target of rapamycin (mTOR) inhibition have yielded disappointing results in glioblastoma (GBM). A major mechanism of resistance involves the activation of a salvage pathway stimulating internal ribosome entry site (IRES)-mediated protein synthesis. PRMT5 activity has been implicated in the enhancement of IRES activity.
Methods: We analyzed the expression and activity of PRMT5 in response to mTOR inhibition in GBM cell lines and short-term patient cultures. To determine whether PRMT5 conferred resistance we used genetic and pharmacological approaches to ablate PRMT5 activity and assessed the effects on in vitro and in vivo sensitivity. Mutational analyses of the requisite IRES-trans-acting factor (ITAF), hnRNP A1 determined whether PRMT5-mediated methylation was necessary for ITAF RNA binding and IRES activity.
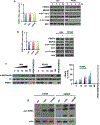
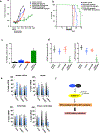
Results: PRMT5 activity is stimulated in response to mTOR inhibitors. Knockdown or treatment with a PRMT5 inhibitor blocked IRES activity and sensitizes GBM cells. Ectopic expression of non-methylatable hnRNP A1 mutants demonstrated that methylation of either arginine residues 218 or 225 was sufficient to maintain IRES binding and hnRNP A1-dependent cyclin D1 or c-MYC IRES activity, however a double R218K/R225K mutant was unable to do so. The PRMT5 inhibitor EPZ015666 displayed synergistic anti-GBM effects in vitro and in a xenograft mouse model in combination with PP242.
Conclusions: These results demonstrate that PRMT5 activity is stimulated upon mTOR inhibition in GBM. Our data further support a signaling cascade in which PRMT5-mediated methylation of hnRNP A1 promotes IRES RNA binding and activation of IRES-mediated protein synthesis and resultant mTOR inhibitor resistance.
Keywords: Drug resistance; EPZ015666; Glioblastoma; PP242; PRMT5; Rapamycin; mTOR.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L, Network TR (2013) The somatic genomic landscape of glioblastoma. Cell 155: 462–477 doi: 10.1016/j.cell.2013.09.034 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

