Mitochondrial Damage and Activation of the STING Pathway Lead to Renal Inflammation and Fibrosis
- PMID: 31474566
- PMCID: PMC7054893
- DOI: 10.1016/j.cmet.2019.08.003
Mitochondrial Damage and Activation of the STING Pathway Lead to Renal Inflammation and Fibrosis
Abstract
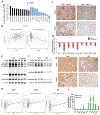
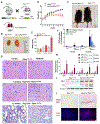
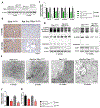
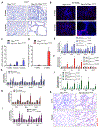
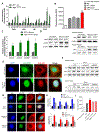
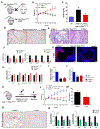
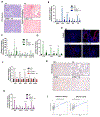
Fibrosis is the final common pathway leading to end-stage renal failure. By analyzing the kidneys of patients and animal models with fibrosis, we observed a significant mitochondrial defect, including the loss of the mitochondrial transcription factor A (TFAM) in kidney tubule cells. Here, we generated mice with tubule-specific deletion of TFAM (Ksp-Cre/Tfamflox/flox). While these mice developed severe mitochondrial loss and energetic deficit by 6 weeks of age, kidney fibrosis, immune cell infiltration, and progressive azotemia causing death were only observed around 12 weeks of age. In renal cells of TFAM KO (knockout) mice, aberrant packaging of the mitochondrial DNA (mtDNA) resulted in its cytosolic translocation, activation of the cytosolic cGAS-stimulator of interferon genes (STING) DNA sensing pathway, and thus cytokine expression and immune cell recruitment. Ablation of STING ameliorated kidney fibrosis in mouse models of chronic kidney disease, demonstrating how TFAM sequesters mtDNA to limit the inflammation leading to fibrosis.
Keywords: TFAM; cGAS-STING pathway; chronic kidney disease; innate immunity; mitochondrial DNA; mitochondrial transcription factor A; renal fibrosis.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures
Comment in
-
STING activation by cytoplasmic mtDNA triggers renal inflammation and fibrosis.Nat Rev Nephrol. 2019 Nov;15(11):661. doi: 10.1038/s41581-019-0211-y. Nat Rev Nephrol. 2019. PMID: 31527791 No abstract available.
References
-
- Bonnet F, and Scheen AJ (2018). Effects of SGLT2 inhibitors on systemic and tissue low-grade inflammation: The potential contribution to diabetes complications and cardiovascular disease. Diabetes Metab 44, 457–464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

