N6-Methyladenosine Modification Controls Circular RNA Immunity
- PMID: 31474572
- PMCID: PMC6778039
- DOI: 10.1016/j.molcel.2019.07.016
N6-Methyladenosine Modification Controls Circular RNA Immunity
Abstract
Circular RNAs (circRNAs) are prevalent in eukaryotic cells and viral genomes. Mammalian cells possess innate immunity to detect foreign circRNAs, but the molecular basis of self versus foreign identity in circRNA immunity is unknown. Here, we show that N6-methyladenosine (m6A) RNA modification on human circRNAs inhibits innate immunity. Foreign circRNAs are potent adjuvants to induce antigen-specific T cell activation, antibody production, and anti-tumor immunity in vivo, and m6A modification abrogates immune gene activation and adjuvant activity. m6A reader YTHDF2 sequesters m6A-circRNA and is essential for suppression of innate immunity. Unmodified circRNA, but not m6A-modified circRNA, directly activates RNA pattern recognition receptor RIG-I in the presence of lysine-63-linked polyubiquitin chain to cause filamentation of the adaptor protein MAVS and activation of the downstream transcription factor IRF3. CircRNA immunity has considerable parallel to prokaryotic DNA restriction modification system that transforms nucleic acid chemical modification into organismal innate immunity.
Keywords: N6-methyladenosine; RIG-I; YTHDF; cancer immunotherapy; circular RNA; self/non-self; vaccine.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
Stanford University and Emory University have filed patent applications based on this work, on which H.Y.C., Y.G.C., R.C., L.A., B.P. and S.K. are named as co-inventors. H.Y.C. is a co-founder and advisor of Accent Therapeutics and Pretzel Therapeutics. H.Y.C. is an advisor of 10X Genomics, Arsenal Biosciences, and Spring Discovery.
Figures
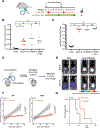

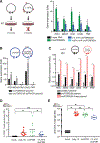



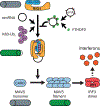
Comment in
-
Going in Circles: The Black Box of Circular RNA Immunogenicity.Mol Cell. 2019 Oct 3;76(1):3-5. doi: 10.1016/j.molcel.2019.08.027. Mol Cell. 2019. PMID: 31585102
References
-
- Baron-Benhamou J, Gehring NH, Kulozik AE, and Hentze MW (2004). Using the λN Peptide to Tether Proteins to RNAs. In mRNA Processing and Metabolism: Methods and Protocols, Schoenberg DR, ed. (Totowa, NJ: Humana Press; ), pp. 135–153. - PubMed
-
- Budhu S, Loike JD, Pandolfi A, Han S, Catalano G, Constantinescu A, Clynes R, and Silverstein SC (2010). CD8+ T cell concentration determines their efficiency in killing cognate antigen–expressing syngeneic mammalian cells in vitro and in mouse tissues. The Journal of Experimental Medicine 207, 223–235. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

