Complete Genome Sequence of Sequevar 14M Ralstonia solanacearum Strain HA4-1 Reveals Novel Type III Effectors Acquired Through Horizontal Gene Transfer
- PMID: 31474968
- PMCID: PMC6703095
- DOI: 10.3389/fmicb.2019.01893
Complete Genome Sequence of Sequevar 14M Ralstonia solanacearum Strain HA4-1 Reveals Novel Type III Effectors Acquired Through Horizontal Gene Transfer
Abstract
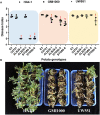
Ralstonia solanacearum, which causes bacterial wilt in a broad range of plants, is considered a "species complex" due to its significant genetic diversity. Recently, we have isolated a new R. solanacearum strain HA4-1 from Hong'an county in Hubei province of China and identified it being phylotype I, sequevar 14M (phylotype I-14M). Interestingly, we found that it can cause various disease symptoms among different potato genotypes and display different pathogenic behavior compared to a phylogenetically related strain, GMI1000. To dissect the pathogenic mechanisms of HA4-1, we sequenced its whole genome by combined sequencing technologies including Illumina HiSeq2000, PacBio RS II, and BAC-end sequencing. Genome assembly results revealed the presence of a conventional chromosome, a megaplasmid as well as a 143 kb plasmid in HA4-1. Comparative genome analysis between HA4-1 and GMI1000 shows high conservation of the general virulence factors such as secretion systems, motility, exopolysaccharides (EPS), and key regulatory factors, but significant variation in the repertoire and structure of type III effectors, which could be the determinants of their differential pathogenesis in certain potato species or genotypes. We have identified two novel type III effectors that were probably acquired through horizontal gene transfer (HGT). These novel R. solanacearum effectors display homology to several YopJ and XopAC family members. We named them as RipBR and RipBS. Notably, the copy of RipBR on the plasmid is a pseudogene, while the other on the megaplasmid is normal. For RipBS, there are three copies located in the megaplasmid and plasmid, respectively. Our results have not only enriched the genome information on R. solanacearum species complex by sequencing the first sequevar 14M strain and the largest plasmid reported in R. solanacearum to date but also revealed the variation in the repertoire of type III effectors. This will greatly contribute to the future studies on the pathogenic evolution, host adaptation, and interaction between R. solanacearum and potato.
Keywords: Ralstonia solanacearum; genome sequencing; plasmid; potato; type III effectors.
Figures

References
-
- Al-Saadi A., Reddy J. D., Duan Y. P., Brunings A. M., Yuan Q., Gabriel D. W. (2007). All five host-range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host-range variation. Mol. Plant Microbe Interact. 20 934–943. 10.1094/MPMI-20-8-0934 - DOI - PubMed
-
- Bertolla F., Frostegård Å, Brito B., Nesme X., Simonet P. (1999). During infection of its host, the plant pathogen Ralstonia solanacearum naturally develops a state of competence and exchanges genetic material. Mol. Plant Microbe Interact. 12 467–472. 10.1094/MPMI.1999.12.5.467 - DOI
-
- Boucher C. A., Barberis P. A., Demery D. A. (1985). Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. Microbiology 131 2449–2457. 10.1099/00221287-131-9-2449 - DOI
LinkOut - more resources
Full Text Sources

