Blimp-1 Contributes to the Development and Function of Regulatory B Cells
- PMID: 31474988
- PMCID: PMC6702260
- DOI: 10.3389/fimmu.2019.01909
Blimp-1 Contributes to the Development and Function of Regulatory B Cells
Abstract
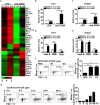
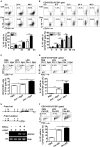
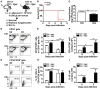
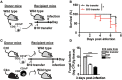
Regulatory B cells (Bregs) are a B cell subset that plays a suppressive role in immune responses. The CD19+CD1dhiCD5+ Bregs that can execute regulatory functions via secreting IL-10 are defined as B10 cells. Bregs suppress autoimmune and inflammatory diseases, whereas they exacerbate infectious diseases caused by bacteria, viruses, or parasites. Notably, the molecular mechanisms regulating the development and functions of Bregs are still largely unknown. Furthermore, the biological impact of Bregs in fungal infection has not yet been demonstrated. Here, we compared the gene expression profiles of IL-10-producing and -non-producing mouse splenic B cells stimulated with lipopolysaccharide (LPS) or anti-CD40 antibody. Blimp-1, a transcription factor known to be critical for plasma cell differentiation, was found to be enriched in the IL-10-producing B cells. The frequency of Blimp-1+ B10 cells was increased in LPS-treated mice and in isolated B10 cells that were stimulated with LPS. Surprisingly, B cell-specific Blimp-1 knockout (Cko) mice, generated by CD19 promoter driven Cre recombinase-dependent deletion of Prdm1 (gene encoding Blimp-1), showed higher frequencies of B10 cells both in the steady state and following injection with LPS, as compared with control littermates. However, B10 cells lacking Blimp-1 failed to efficiently suppress the proliferation of naïve CD4+ T cells primed with anti-CD3 and anti-CD28 antibodies. B10 cells can be stimulated for further differentiation into plasmablasts, and a subset of plasmablasts express IL-10. We found that B10 cells from Cko mice failed to generate both IL-10-non-producing and IL-10-producing plasmablasts. Mechanistically, we found that Blimp-1 can directly suppress Il-10, whereas, in the presence of activated STAT3, Blimp-1 works together with activated STAT3 to upregulate Il-10. Moreover, we also found that B10 cells improve the clearance of Candida albicans infection but worsen the infection mortality. Notably, a lack of Blimp-1 in B10 cells did not change these effects of adoptively transferred B10 cells on fungal infections. Together, our data show that Blimp-1 regulates the generation, differentiation, and IL-10 production of Bregs.
Keywords: B10 cells; Blimp-1; Candida albicans; IL-10; regulatory B cells.
Figures


Similar articles
-
Astilbin combined with lipopolysaccharide induces IL-10-producing regulatory B cells via the STAT3 signalling pathway.Biomed Pharmacother. 2020 Sep;129:110450. doi: 10.1016/j.biopha.2020.110450. Epub 2020 Jul 7. Biomed Pharmacother. 2020. PMID: 32768945
-
TLR4 supports the expansion of FasL+CD5+CD1dhi regulatory B cells, which decreases in contact hypersensitivity.Mol Immunol. 2017 Jul;87:188-199. doi: 10.1016/j.molimm.2017.04.016. Epub 2017 May 12. Mol Immunol. 2017. PMID: 28505514
-
Regulatory B and T cell responses in patients with autoimmune thyroid disease and healthy controls.Dan Med J. 2016 Feb;63(2):B5177. Dan Med J. 2016. PMID: 26836805 Review.
-
Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions.Nature. 2012 Nov 8;491(7423):264-8. doi: 10.1038/nature11501. Epub 2012 Oct 14. Nature. 2012. PMID: 23064231 Free PMC article.
-
The role of B regulatory (B10) cells in inflammatory disorders and their potential as therapeutic targets.Int Immunopharmacol. 2020 Jan;78:106111. doi: 10.1016/j.intimp.2019.106111. Epub 2019 Dec 24. Int Immunopharmacol. 2020. PMID: 31881524 Review.
Cited by
-
Properties of regulatory B cells regulating B cell targets.Am J Transplant. 2021 Dec;21(12):3847-3857. doi: 10.1111/ajt.16772. Epub 2021 Sep 6. Am J Transplant. 2021. PMID: 34327838 Free PMC article.
-
Molecular Mechanisms Driving IL-10- Producing B Cells Functions: STAT3 and c-MAF as Underestimated Central Key Regulators?Front Immunol. 2022 Mar 10;13:818814. doi: 10.3389/fimmu.2022.818814. eCollection 2022. Front Immunol. 2022. PMID: 35359922 Free PMC article.
-
The trehalose glycolipid C18Brar promotes antibody and T-cell immune responses to Mannheimia haemolytica and Mycoplasma ovipneumoniae whole cell antigens in sheep.PLoS One. 2023 Jan 19;18(1):e0278853. doi: 10.1371/journal.pone.0278853. eCollection 2023. PLoS One. 2023. PMID: 36656850 Free PMC article.
-
Schistosoma japonicum-derived peptide SJMHE1 ameliorates allergic symptoms and responses in mice with allergic rhinitis.Front Cell Infect Microbiol. 2023 Jun 6;13:1143950. doi: 10.3389/fcimb.2023.1143950. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37346033 Free PMC article.
-
Expansion of B10 cells in vitro: Pathways, techniques and applications in transplantation (Review).Int J Mol Med. 2025 Feb;55(2):29. doi: 10.3892/ijmm.2024.5470. Epub 2024 Dec 13. Int J Mol Med. 2025. PMID: 39670296 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

