The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K
- PMID: 31477691
- PMCID: PMC6718402
- DOI: 10.1038/s41467-019-11753-1
The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K
Abstract
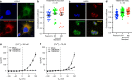
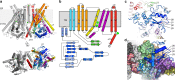
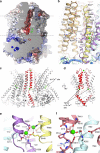
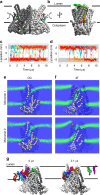
Membranes in cells have defined distributions of lipids in each leaflet, controlled by lipid scramblases and flip/floppases. However, for some intracellular membranes such as the endoplasmic reticulum (ER) the scramblases have not been identified. Members of the TMEM16 family have either lipid scramblase or chloride channel activity. Although TMEM16K is widely distributed and associated with the neurological disorder autosomal recessive spinocerebellar ataxia type 10 (SCAR10), its location in cells, function and structure are largely uncharacterised. Here we show that TMEM16K is an ER-resident lipid scramblase with a requirement for short chain lipids and calcium for robust activity. Crystal structures of TMEM16K show a scramblase fold, with an open lipid transporting groove. Additional cryo-EM structures reveal extensive conformational changes from the cytoplasmic to the ER side of the membrane, giving a state with a closed lipid permeation pathway. Molecular dynamics simulations showed that the open-groove conformation is necessary for scramblase activity.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- BB/P01948X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I019855/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- P41 GM103311/GM/NIGMS NIH HHS/United States
- FS/17/45/33102/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

