mRNA structure determines modification by pseudouridine synthase 1
- PMID: 31477916
- PMCID: PMC6764900
- DOI: 10.1038/s41589-019-0353-z
mRNA structure determines modification by pseudouridine synthase 1
Abstract
Pseudouridine (Ψ) is a post-transcriptional RNA modification that alters RNA-RNA and RNA-protein interactions that affect gene expression. Messenger RNA pseudouridylation was recently discovered as a widespread and conserved phenomenon, but the mechanisms responsible for selective, regulated pseudouridylation of specific sequences within mRNAs were unknown. Here, we have revealed mRNA targets for five pseudouridine synthases and probed the determinants of mRNA target recognition by the predominant mRNA pseudouridylating enzyme, Pus1, by developing high-throughput kinetic analysis of pseudouridylation in vitro. Combining computational prediction and rational mutational analysis revealed an RNA structural motif that is both necessary and sufficient for mRNA pseudouridylation. Applying this structural context information predicted hundreds of additional mRNA targets that were pseudouridylated in vivo. These results demonstrate a structure-dependent mode of mRNA target recognition by a conserved pseudouridine synthase and implicate modulation of RNA structure as the probable mechanism to regulate mRNA pseudouridylation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
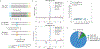

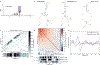
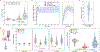


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

