Follicular regulatory T cells control humoral and allergic immunity by restraining early B cell responses
- PMID: 31477921
- PMCID: PMC6754271
- DOI: 10.1038/s41590-019-0472-4
Follicular regulatory T cells control humoral and allergic immunity by restraining early B cell responses
Abstract
Follicular regulatory T (TFR) cells have specialized roles in modulating follicular helper T (TFH) cell activation of B cells. However, the precise role of TFR cells in controlling antibody responses to foreign antigens and autoantigens in vivo is still unclear due to a lack of specific tools. A TFR cell-deleter mouse was developed that selectively deletes TFR cells, facilitating temporal studies. TFR cells were found to regulate early, but not late, germinal center (GC) responses to control antigen-specific antibody and B cell memory. Deletion of TFR cells also resulted in increased self-reactive immunoglobulin (Ig) G and IgE. The increased IgE levels led us to interrogate the role of TFR cells in house dust mite models. TFR cells were found to control TFH13 cell-induced IgE. In vivo, loss of TFR cells increased house-dust-mite-specific IgE and lung inflammation. Thus, TFR cells control IgG and IgE responses to vaccines, allergens and autoantigens, and exert critical immunoregulatory functions before GC formation.
Conflict of interest statement
Competing interests
The authors declare no competing interests
Figures



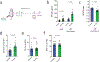


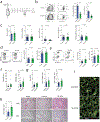
References
-
- Crotty S Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29, 621–663 (2011). - PubMed
-
- Victora GD & Nussenzweig MC Germinal centers. Annu Rev Immunol 30, 429–457 (2012). - PubMed
-
- Vinuesa CG, Linterman MA, Yu D & MacLennan IC Follicular Helper T Cells. Annu Rev Immunol 34, 335–368 (2016). - PubMed
Methods only References
-
- Bettelli E et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006). - PubMed
-
- Sage PT & Sharpe AH In Vitro Assay to Sensitively Measure TFR Suppressive Capacity and TFH Stimulation of B Cell Responses. Methods Mol Biol 1291, 151–160 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

