Interferon lambda 4 impacts the genetic diversity of hepatitis C virus
- PMID: 31478835
- PMCID: PMC6721795
- DOI: 10.7554/eLife.42463
Interferon lambda 4 impacts the genetic diversity of hepatitis C virus
Abstract
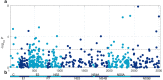
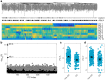
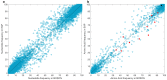
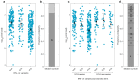
Hepatitis C virus (HCV) is a highly variable pathogen that frequently establishes chronic infection. This genetic variability is affected by the adaptive immune response but the contribution of other host factors is unclear. Here, we examined the role played by interferon lambda-4 (IFN-λ4) on HCV diversity; IFN-λ4 plays a crucial role in spontaneous clearance or establishment of chronicity following acute infection. We performed viral genome-wide association studies using human and viral data from 485 patients of white ancestry infected with HCV genotype 3a. We demonstrate that combinations of host genetic variants, which determine IFN-λ4 protein production and activity, influence amino acid variation across the viral polyprotein - not restricted to specific viral proteins or HLA restricted epitopes - and modulate viral load. We also observed an association with viral di-nucleotide proportions. These results support a direct role for IFN-λ4 in exerting selective pressure across the viral genome, possibly by a novel mechanism.
Keywords: genetics; genome-to-genome analysis; genomics; hepatitis C virus; host-pathogen interactions; human; infectious disease; innate immunity; interferon lambda 4; microbiology; virus.
© 2019, Ansari et al.
Conflict of interest statement
MA, EA, CI, Ad, SL, CB, DB, AT, PP, VS, VC, EH, RB, AP, ET, PS, PK, CH, EB, CS, JM, VP No competing interests declared, GF Grants Consulting and Speaker/Advisory Board: AbbVie, Alcura, Bristol-Myers Squibb, Gilead, Janssen, GlaxoSmithKline, Merck, Roche, Springbank, Idenix, Tekmira, Novartis, WI Grants, Consulting and Advisory/ Speaker Board: Roche, Janssen Cilag, Gilead Sciences, Novartis, GlaxoSmithKline, Pfizer, Abbvie and Bristol-Myers Squibb, KA Grants, Consulting and Advisory/ Speaker Board: Achillion, Alnylam, Astellas, Abbvie, Bristol-Myers Squibb, Gilead, GlaxoSmithKline, Janssen, Merck, Roche, Novartis, Vir
Figures







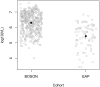

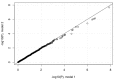
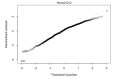
Comment in
-
What makes the hepatitis C virus evolve?Elife. 2019 Sep 3;8:e50148. doi: 10.7554/eLife.50148. Elife. 2019. PMID: 31478837 Free PMC article.
References
-
- Angulo J, Pino K, Echeverría-Chagas N, Marco C, Martínez-Valdebenito C, Galeno H, Villagra E, Vera L, Lagos N, Becerra N, Mora J, Bermúdez A, Cárcamo M, Díaz J, Miquel JF, Ferrés M, López-Lastra M. Association of Single-Nucleotide polymorphisms in IL28B, but not TNF-α, with severity of disease caused by andes virus. Clinical Infectious Diseases : An Official Publication of the Infectious Diseases Society of America. 2015;61:e62–e69. doi: 10.1093/cid/civ830. - DOI - PMC - PubMed
-
- Ansari MA, Pedergnana V, L C Ip C, Magri A, Von Delft A, Bonsall D, Chaturvedi N, Bartha I, Smith D, Nicholson G, McVean G, Trebes A, Piazza P, Fellay J, Cooke G, Foster GR, Hudson E, McLauchlan J, Simmonds P, Bowden R, Klenerman P, Barnes E, Spencer CCA, STOP-HCV Consortium Genome-to-genome analysis highlights the effect of the human innate and adaptive immune systems on the hepatitis C virus. Nature Genetics. 2017;49:666–673. doi: 10.1038/ng.3835. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 097364/Z/11/Z/Wellcome/International
- MC_UU_12014/1/MRC_/Medical Research Council/United Kingdom
- MR/K01532X/1 - STOP-HCV Consortium/MRC_/Medical Research Council/United Kingdom
- RP-2016-07-012/DH_/Department of Health/United Kingdom
- 14/02/17/DH_/Department of Health/United Kingdom
- C0365/MRF_/MRF_/United Kingdom
- MC_PC_14131/MRC_/Medical Research Council/United Kingdom
- MR/N005953/1/MRC_/Medical Research Council/United Kingdom
- 090532/Z/09/Z/Wellcome/International
- MC_UU 12014/2/MRC_/Medical Research Council/United Kingdom
- 109965MA/Oxford Martin School, University of Oxford/International
- MR/K01532X/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12014/2/MRC_/Medical Research Council/United Kingdom
- MC_U130184143/MRC_/Medical Research Council/United Kingdom
- 109965/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- U19 AI082630/AI/NIAID NIH HHS/United States
- MC_UU_12014/12/MRC_/Medical Research Council/United Kingdom
- 102789/Z/13/Z/Wellcome/International
- U19AI082630/National Institute for Health Research/International
LinkOut - more resources
Full Text Sources
Medical
Research Materials

