Structures of the substrate-binding protein YfeA in apo and zinc-reconstituted holo forms
- PMID: 31478906
- PMCID: PMC6719664
- DOI: 10.1107/S2059798319010866
Structures of the substrate-binding protein YfeA in apo and zinc-reconstituted holo forms
Abstract
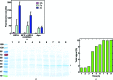
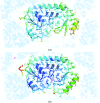
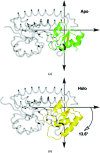
In the structural biology of bacterial substrate-binding proteins (SBPs), a growing number of comparisons between substrate-bound and substrate-free forms of metal atom-binding (cluster A-I) SBPs have revealed minimal structural differences between forms. These observations contrast with SBPs that bind substrates such as amino acids or nucleic acids and may undergo >60° rigid-body rotations. Substrate transfer in these SBPs is described by a Venus flytrap model, although this model may not apply to all SBPs. In this report, structures are presented of substrate-free (apo) and reconstituted substrate-bound (holo) YfeA, a polyspecific cluster A-I SBP from Yersinia pestis. It is demonstrated that an apo cluster A-I SBP can be purified by fractionation when co-expressed with its cognate transporter, adding an alternative strategy to the mutagenesis or biochemical treatment used to generate other apo cluster A-I SBPs. The apo YfeA structure contains 111 disordered protein atoms in a mobile helix located in the flexible carboxy-terminal lobe. Metal binding triggers a 15-fold reduction in the solvent-accessible surface area of the metal-binding site and reordering of the 111 protein atoms in the mobile helix. The flexible lobe undergoes a 13.6° rigid-body rotation that is driven by a spring-hammer metal-binding mechanism. This asymmetric rigid-body rotation may be unique to metal atom-binding SBPs (i.e. clusters A-I, A-II and D-IV).
Keywords: X-ray crystallography; Yersinia pestis; YfeA; cluster A-I; substrate transfer; substrate-binding protein.
open access.
Figures



References
-
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. - PMC - PubMed
-
- Ahuja, S., Rougé, L., Swem, D. L., Sudhamsu, J., Wu, P., Russell, S. J., Alexander, M. K., Tam, C., Nishiyama, M., Starovasnik, M. A. & Koth, C. M. (2015). Structure, 23, 713–723. - PubMed
-
- Bearden, S. W. & Perry, R. D. (1999). Mol. Microbiol. 32, 403–414. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

