A preliminary study of microRNA expression in different types of primary melanoma
- PMID: 31479413
- PMCID: PMC7202197
- DOI: 10.17305/bjbms.2019.4271
A preliminary study of microRNA expression in different types of primary melanoma
Abstract
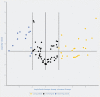
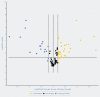
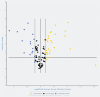
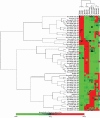
MicroRNAs (miRNAs) have been proven to regulate the development and progression of cancer through various mechanisms. The aim of the present study was to compare miRNA expression between primary melanomas from different sites. We analyzed the expression of 84 miRNAs in 27 primary melanoma and 5 nevus formalin-fixed paraffin-embedded (FFPE) samples using the Human Cancer PathwayFinder miScript miRNA PCR Array. The FFPE samples were obtained from the archives of the Municipal Clinical Emergency Hospital of Timisoara and included 10 cutaneous melanomas, 10 uveal melanomas, 7 mucosal melanomas, and 5 cutaneous nevi. Out of 84 miRNAs, 11 miRNAs showed altered expression in all types of melanoma compared with the nevi. Among these, miR-155-5p, miR-9-5p, miR-142-5p, miR-19a-3p, miR-134-5p, and miR-301a-3p were upregulated, while miR-205-5p, miR-203a-3p, miR-27b-3p, miR-218-5p, and miR-23b-3p were downregulated. The highest similarity in miRNA expression pattern was found between uveal and mucosal melanoma groups, i.e., 15 miRNAs had altered expression in both groups. Overall, we identified several miRNAs with significantly altered expression in primary melanomas, including those reported for the first time in this type of cancer. Among them, mir-9-5p, mir-203a-3p, mir-19a-3p, mir-27b-3p, and mir-218-5p showed altered expression in all three melanoma types vs. nevi. Further research should explore the potential of these miRNAs in melanoma.
Conflict of interest statement
Conflict of interest statement: The authors declare no conflict of interests
Figures
References
-
- Ocanha-Xavier JP, Xavier-Junior JCC, Marques MEA. Melanoma:clinical, evolutive and histopathological characteristics of a series of 136 cases. An Bras Dermatol. 2018;93(3):373–6. https://doi.org/10.1590/abd1806-4841.201⇝0. - PMC - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics 2016. CA Cancer J Clin. 2016;66(1):7–30. https://doi.org/10.3322/caac.21332. - PubMed
-
- Fechete O, Ungureanu L, Şenilă S, Vornicescu D, Dănescu S, Vasilovici A, et al. Risk factors for melanoma and skin health behaviour:an analysis on Romanian melanoma patients. Oncol Lett. 2019;17(5):4139–44. https://doi.org/10.3892/ol.2018.9737. - PMC - PubMed
-
- Barrett AW, Raja AM. The immunohistochemical identification of human oral mucosal melanocytes. Arch Oral Biol. 1997;42(1):77–81. https://doi.org/10.1016/S0003-9969(96)00113-6. - PubMed
-
- Tolleson WH. Human melanocyte biology, toxicology, and pathology. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2005;23(2):105–61. https://doi.org/10.1080/10590500500234970. - PubMed

