Fetal exposure to the maternal microbiota in humans and mice
- PMID: 31479427
- PMCID: PMC6795398
- DOI: 10.1172/jci.insight.127806
Fetal exposure to the maternal microbiota in humans and mice
Abstract
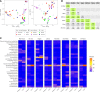
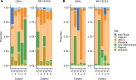
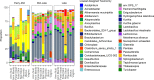
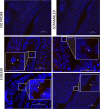
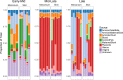
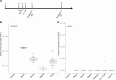
Previous studies have demonstrated the presence of microbial DNA in the fetal environment. However, it remains unclear whether this DNA represents viable bacteria and how it relates to the maternal microbiota across body sites. We studied the microbiota of human and mouse dyads to understand these relationships, localize bacteria in the fetus, and demonstrate bacterial viability. In human preterm and full-term mother-infant dyads at the time of cesarean delivery, the oral cavity and meconium of newborn infants born as early as 24 weeks of gestation contained a microbiota that was predicted to originate from in utero sources, including the placenta. Using operative deliveries of pregnant mice under highly controlled, sterile conditions in the laboratory, composition, visualization, and viability of bacteria in the in utero compartment and fetal intestine were demonstrated by 16S rRNA gene sequencing, fluorescence in situ hybridization, and bacterial culture. The composition and predicted source of the fetal gut microbiota shifted between mid- and late gestation. Cultivatable bacteria in the fetal intestine were found during mid-gestation but not late gestation. Our results demonstrate a dynamic, viable mammalian fetal microbiota during in utero development.
Keywords: Bacterial infections; Development; Microbiology.
Conflict of interest statement
Figures

Similar articles
-
No Consistent Evidence for Microbiota in Murine Placental and Fetal Tissues.mSphere. 2020 Feb 26;5(1):e00933-19. doi: 10.1128/mSphere.00933-19. mSphere. 2020. PMID: 32102944 Free PMC article.
-
Viable bacterial colonization is highly limited in the human intestine in utero.Nat Med. 2020 Apr;26(4):599-607. doi: 10.1038/s41591-020-0761-3. Epub 2020 Feb 24. Nat Med. 2020. PMID: 32094926 Free PMC article.
-
Presence of distinctive microbiome in the first-pass meconium of newborn infants.Sci Rep. 2021 Sep 30;11(1):19449. doi: 10.1038/s41598-021-98951-4. Sci Rep. 2021. PMID: 34593932 Free PMC article.
-
Planting the seed: Origins, composition, and postnatal health significance of the fetal gastrointestinal microbiota.Crit Rev Microbiol. 2017 May;43(3):352-369. doi: 10.1080/1040841X.2016.1211088. Epub 2016 Dec 8. Crit Rev Microbiol. 2017. PMID: 27931152 Review.
-
Investigating prenatal and perinatal factors on meconium microbiota: a systematic review and cohort study.Pediatr Res. 2024 Jan;95(1):135-145. doi: 10.1038/s41390-023-02783-z. Epub 2023 Aug 17. Pediatr Res. 2024. PMID: 37591927 Free PMC article.
Cited by
-
The Maternal-Fetal Gut Microbiota Axis: Physiological Changes, Dietary Influence, and Modulation Possibilities.Life (Basel). 2022 Mar 15;12(3):424. doi: 10.3390/life12030424. Life (Basel). 2022. PMID: 35330175 Free PMC article. Review.
-
Does the Amniotic Fluid of Mice Contain a Viable Microbiota?Front Immunol. 2022 Feb 28;13:820366. doi: 10.3389/fimmu.2022.820366. eCollection 2022. Front Immunol. 2022. PMID: 35296083 Free PMC article.
-
Human Fetal Lungs Harbor a Microbiome Signature.Am J Respir Crit Care Med. 2020 Apr 15;201(8):1002-1006. doi: 10.1164/rccm.201911-2127LE. Am J Respir Crit Care Med. 2020. PMID: 31898918 Free PMC article. No abstract available.
-
Impacts of Delivery Mode and Maternal Factors on Neonatal Oral Microbiota.Front Microbiol. 2022 Jun 27;13:915423. doi: 10.3389/fmicb.2022.915423. eCollection 2022. Front Microbiol. 2022. PMID: 35832807 Free PMC article.
-
The Role of Intestinal Bacteria and Gut-Brain Axis in Hepatic Encephalopathy.Front Cell Infect Microbiol. 2021 Jan 21;10:595759. doi: 10.3389/fcimb.2020.595759. eCollection 2020. Front Cell Infect Microbiol. 2021. PMID: 33553004 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

