Peptide-conjugated oligonucleotides evoke long-lasting myotonic dystrophy correction in patient-derived cells and mice
- PMID: 31479430
- PMCID: PMC6819114
- DOI: 10.1172/JCI128205
Peptide-conjugated oligonucleotides evoke long-lasting myotonic dystrophy correction in patient-derived cells and mice
Abstract
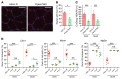
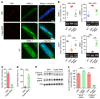
Antisense oligonucleotides (ASOs) targeting pathologic RNAs have shown promising therapeutic corrections for many genetic diseases including myotonic dystrophy (DM1). Thus, ASO strategies for DM1 can abolish the toxic RNA gain-of-function mechanism caused by nucleus-retained mutant DMPK (DM1 protein kinase) transcripts containing CUG expansions (CUGexps). However, systemic use of ASOs for this muscular disease remains challenging due to poor drug distribution to skeletal muscle. To overcome this limitation, we test an arginine-rich Pip6a cell-penetrating peptide and show that Pip6a-conjugated morpholino phosphorodiamidate oligomer (PMO) dramatically enhanced ASO delivery into striated muscles of DM1 mice following systemic administration in comparison with unconjugated PMO and other ASO strategies. Thus, low-dose treatment with Pip6a-PMO-CAG targeting pathologic expansions is sufficient to reverse both splicing defects and myotonia in DM1 mice and normalizes the overall disease transcriptome. Moreover, treated DM1 patient-derived muscle cells showed that Pip6a-PMO-CAG specifically targets mutant CUGexp-DMPK transcripts to abrogate the detrimental sequestration of MBNL1 splicing factor by nuclear RNA foci and consequently MBNL1 functional loss, responsible for splicing defects and muscle dysfunction. Our results demonstrate that Pip6a-PMO-CAG induces long-lasting correction with high efficacy of DM1-associated phenotypes at both molecular and functional levels, and strongly support the use of advanced peptide conjugates for systemic corrective therapy in DM1.
Keywords: Genetic diseases; Muscle; Therapeutics.
Conflict of interest statement
Figures


Comment in
-
Better living through peptide-conjugated chemistry: next-generation antisense oligonucleotides.J Clin Invest. 2019 Nov 1;129(11):4570-4571. doi: 10.1172/JCI131933. J Clin Invest. 2019. PMID: 31566581 Free PMC article.
References
-
- Harper PS. Myotonic Dystrophy. London, UK: Oxford University Press; 2001.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

