Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome
- PMID: 31481669
- PMCID: PMC6722078
- DOI: 10.1038/s41467-019-11951-x
Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome
Abstract
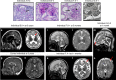
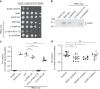
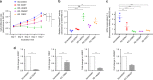
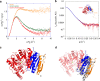
N6-threonyl-carbamoylation of adenosine 37 of ANN-type tRNAs (t6A) is a universal modification essential for translational accuracy and efficiency. The t6A pathway uses two sequentially acting enzymes, YRDC and OSGEP, the latter being a subunit of the multiprotein KEOPS complex. We recently identified mutations in genes encoding four out of the five KEOPS subunits in children with Galloway-Mowat syndrome (GAMOS), a clinically heterogeneous autosomal recessive disease characterized by early-onset steroid-resistant nephrotic syndrome and microcephaly. Here we show that mutations in YRDC cause an extremely severe form of GAMOS whereas mutations in GON7, encoding the fifth KEOPS subunit, lead to a milder form of the disease. The crystal structure of the GON7/LAGE3/OSGEP subcomplex shows that the intrinsically disordered GON7 protein becomes partially structured upon binding to LAGE3. The structure and cellular characterization of GON7 suggest its involvement in the cellular stability and quaternary arrangement of the KEOPS complex.
Conflict of interest statement
F.H. is a cofounder and SAB member of Goldfinch-Bio. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

