Expression of immune regulatory genes correlate with the abundance of specific Clostridiales and Verrucomicrobia species in the equine ileum and cecum
- PMID: 31481726
- PMCID: PMC6722064
- DOI: 10.1038/s41598-019-49081-5
Expression of immune regulatory genes correlate with the abundance of specific Clostridiales and Verrucomicrobia species in the equine ileum and cecum
Abstract
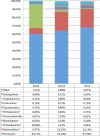
Billions of bacteria inhabit the gastrointestinal tract. Immune-microbial cross talk is responsible for immunological homeostasis, and symbiotic microbial species induce regulatory immunity, which helps to control the inflammation levels. In this study we aimed to identify species within the equine intestinal microbiota with the potential to induce regulatory immunity. These could be future targets for preventing or treating low-grade chronic inflammation occurring as a result of intestinal microbial changes and disruption of the homeostasis. 16S rRNA gene amplicon sequencing was performed on samples of intestinal microbial content from ileum, cecum, and colon of 24 healthy horses obtained from an abattoir. Expression of genes coding for IL-6, IL-10, IL-12, IL-17, 18 s, TNFα, TGFβ, and Foxp3 in the ileum and mesenteric lymph nodes was measured by qPCR. Intestinal microbiota composition was significantly different in the cecum and colon compared to the ileum, which contains large abundances of Proteobacteria. Especially members of the Clostridiales order correlated positively with the regulatory T-cell transcription factor Foxp3 and so did the phylum Verrucomicrobia. We conclude that Clostridiales and Verrucomicrobia have the potential to induce regulatory immunity and are possible targets for intestinal microbial interventions aiming at regulatory immunity improvement.
Conflict of interest statement
The authors declare no conflicts of interest, apart from the fact that F.L. is employed by Brogaarden Diets Aps.
Figures
Similar articles
-
Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments of Mongolian horses.Microbiologyopen. 2020 Jun;9(6):1085-1101. doi: 10.1002/mbo3.1020. Epub 2020 Mar 9. Microbiologyopen. 2020. PMID: 32153142 Free PMC article.
-
A Microbiological Map of the Healthy Equine Gastrointestinal Tract.PLoS One. 2016 Nov 15;11(11):e0166523. doi: 10.1371/journal.pone.0166523. eCollection 2016. PLoS One. 2016. PMID: 27846295 Free PMC article.
-
Intense Exercise and Aerobic Conditioning Associated with Chromium or L-Carnitine Supplementation Modified the Fecal Microbiota of Fillies.PLoS One. 2016 Dec 9;11(12):e0167108. doi: 10.1371/journal.pone.0167108. eCollection 2016. PLoS One. 2016. PMID: 27935992 Free PMC article.
-
Impact of a probiotic, inulin, or their combination on the piglets' microbiota at different intestinal locations.Benef Microbes. 2015;6(4):473-83. doi: 10.3920/BM2014.0030. Epub 2015 Feb 12. Benef Microbes. 2015. PMID: 25380797
-
A high-throughput DNA sequencing study of fecal bacteria of seven Mexican horse breeds.Arch Microbiol. 2022 Jun 10;204(7):382. doi: 10.1007/s00203-022-03009-2. Arch Microbiol. 2022. PMID: 35687150
Cited by
-
Socializing Models During Lactation Alter Colonic Mucosal Gene Expression and Fecal Microbiota of Growing Piglets.Front Microbiol. 2022 Jul 7;13:819011. doi: 10.3389/fmicb.2022.819011. eCollection 2022. Front Microbiol. 2022. PMID: 35875524 Free PMC article.
-
Racial-Ethnic Disparities of Obesity Require Community Context-Specific Biomedical Research for Native Hawaiians and Other Pacific Islanders.Nutrients. 2024 Dec 11;16(24):4268. doi: 10.3390/nu16244268. Nutrients. 2024. PMID: 39770890 Free PMC article. Review.
-
Development of the equine gut microbiota.Sci Rep. 2019 Oct 8;9(1):14427. doi: 10.1038/s41598-019-50563-9. Sci Rep. 2019. PMID: 31594971 Free PMC article.
-
Effect of Broussonetia papyrifera silage on the serum indicators, hindgut parameters and fecal bacterial community of Holstein heifers.AMB Express. 2020 Oct 31;10(1):197. doi: 10.1186/s13568-020-01135-y. AMB Express. 2020. PMID: 33128623 Free PMC article.
-
Role of Gut Microbiota and Metabolite Remodeling on the Development and Management of Rheumatoid Arthritis: A Narrative Review.Vet Sci. 2025 Jul 5;12(7):642. doi: 10.3390/vetsci12070642. Vet Sci. 2025. PMID: 40711302 Free PMC article. Review.
References
-
- Tlaskalová-Hogenová Helena, Štěpánková Renata, Kozáková Hana, Hudcovic Tomáš, Vannucci Luca, Tučková Ludmila, Rossmann Pavel, Hrnčíř Tomáš, Kverka Miloslav, Zákostelská Zuzana, Klimešová Klára, Přibylová Jaroslava, Bártová Jiřina, Sanchez Daniel, Fundová Petra, Borovská Dana, Šrůtková Dagmar, Zídek Zdeněk, Schwarzer Martin, Drastich Pavel, Funda David P. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cellular & Molecular Immunology. 2011;8(2):110–120. doi: 10.1038/cmi.2010.67. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

