Combined poly-ADP ribose polymerase and ataxia-telangiectasia mutated/Rad3-related inhibition targets ataxia-telangiectasia mutated-deficient lung cancer cells
- PMID: 31481733
- PMCID: PMC6889280
- DOI: 10.1038/s41416-019-0565-8
Combined poly-ADP ribose polymerase and ataxia-telangiectasia mutated/Rad3-related inhibition targets ataxia-telangiectasia mutated-deficient lung cancer cells
Erratum in
-
Correction: Combined poly-ADP ribose polymerase and ataxia-telangiectasia mutated/Rad3-related inhibition targets ataxia-telangiectasia mutated-deficient lung cancer cells.Br J Cancer. 2020 Jun;122(12):1872. doi: 10.1038/s41416-020-0841-7. Br J Cancer. 2020. PMID: 32303715 Free PMC article.
Abstract
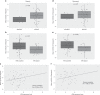
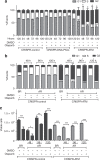
Background: Up to 40% of lung adenocarcinoma have been reported to lack ataxia-telangiectasia mutated (ATM) protein expression. We asked whether ATM-deficient lung cancer cell lines are sensitive to poly-ADP ribose polymerase (PARP) inhibitors and determined the mechanism of action of olaparib in ATM-deficient A549 cells.
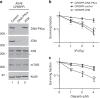
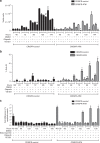
Methods: We analysed drug sensitivity data for olaparib and talazoparib in lung adenocarcinoma cell lines from the Genomics of Drug Sensitivity in Cancer (GDSC) project. We deleted ATM from A549 lung adenocarcinoma cells using CRISPR/Cas9 and determined the effects of olaparib and the ATM/Rad3-related (ATR) inhibitor VE-821 on cell viability.
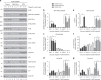
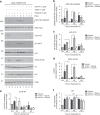
Results: IC50 values for both olaparib and talazoparib positively correlated with ATM mRNA levels and gene amplification status in lung adenocarcinoma cell lines. ATM mutation was associated with a significant decrease in the IC50 for olaparib while a similar trend was observed for talazoparib. A549 cells with deletion of ATM were sensitive to ionising radiation and olaparib. Olaparib induced phosphorylation of DNA damage markers and reversible G2 arrest in ATM-deficient cells, while the combination of olaparib and VE-821 induced cell death.
Conclusions: Patients with tumours characterised by ATM-deficiency may benefit from treatment with a PARP inhibitor in combination with an ATR inhibitor.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

