Action of Dicumarol on Glucosamine-1-Phosphate Acetyltransferase of GlmU and Mycobacterium tuberculosis
- PMID: 31481936
- PMCID: PMC6710349
- DOI: 10.3389/fmicb.2019.01799
Action of Dicumarol on Glucosamine-1-Phosphate Acetyltransferase of GlmU and Mycobacterium tuberculosis
Abstract
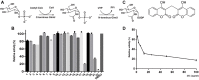
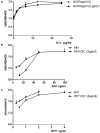
Mycobacterium tuberculosis is one of most pathogenic microorganisms in the world. Previously, the bifunctional enzyme GlmU with glucosamine-1-phosphate acetyltransferase activity and N-acetylglucosamine-1-phosphate uridyltransferase activity has been suggested as a potential drug target; therefore, discovering compounds targeting GlmU acetyltransferase is necessary. The natural products were tested for inhibition of GlmU acetyltransferase activity. We found that dicumarol exhibited inhibitory effects on GlmU acetyltransferase, with a concentration achieving a 50% inhibition (IC50) value of 4.608 μg/ml (13.7 μM). The inhibition kinetics indicated that dicumarol uncompetitively inhibited acetyl CoA and showed mixed-type inhibition for glucosamine-1-phosphate (GlcN-1-P). The activity of dicumarol against M. tuberculosis H37Ra was evaluated with a minimum inhibitory concentration (MIC) value of 6.25 μg/ml (18.55 μM) in the Alamar blue assay. Dicumarol also exhibited inhibitory effects on several clinically sensitive M. tuberculosis strains and drug-resistant strains, with a range of MIC value of 6.25 to >100 μg/ml. Dicumarol increased the sensitivity of anti-tuberculosis drugs (isoniazid and rifampicin) when dicumarol was present at a low concentration. The transcriptome and proteome data of M. tuberculosis H37Ra treated by dicumarol showed that the affected genes were associated with cell wall synthesis, DNA damage and repair, metabolic processes, and signal transduction. These results provided the mechanism of dicumarol inhibition against GlmU acetyltransferase and M. tuberculosis and also suggested that dicumarol is a potential candidate for TB treatment.
Keywords: GlmU; Mycobacterium tuberculosis; acetyltransferase; cell wall; dicumarol; inhibitor.
Figures




Similar articles
-
GlmU inhibitor from the roots of Euphorbia ebracteolata as an anti-tuberculosis agent.RSC Adv. 2022 Jun 22;12(28):18266-18273. doi: 10.1039/d2ra02044k. eCollection 2022 Jun 14. RSC Adv. 2022. PMID: 35800323 Free PMC article.
-
High-throughput screen identifies small molecule inhibitors targeting acetyltransferase activity of Mycobacterium tuberculosis GlmU.Tuberculosis (Edinb). 2015 Dec;95(6):664-677. doi: 10.1016/j.tube.2015.06.003. Epub 2015 Jul 31. Tuberculosis (Edinb). 2015. PMID: 26318557
-
Expression, essentiality, and a microtiter plate assay for mycobacterial GlmU, the bifunctional glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase.Int J Biochem Cell Biol. 2008;40(11):2560-71. doi: 10.1016/j.biocel.2008.05.003. Epub 2008 May 15. Int J Biochem Cell Biol. 2008. PMID: 18573680 Free PMC article.
-
Insights into the central role of N-acetyl-glucosamine-1-phosphate uridyltransferase (GlmU) in peptidoglycan metabolism and its potential as a therapeutic target.Biochem J. 2023 Jun 18;480(15):1147-1164. doi: 10.1042/BCJ20230173. Biochem J. 2023. PMID: 37498748 Review.
-
Mechanism and Potential Inhibitors of GlmU: A Novel Target for Antimicrobial Drug Discovery.Curr Drug Targets. 2017;18(14):1587-1597. doi: 10.2174/1389450117666160502152011. Curr Drug Targets. 2017. PMID: 27138757 Review.
Cited by
-
Dietary supplementation with Neolamarckia cadamba leaf extract improves broiler meat quality by enhancing antioxidant capacity and regulating metabolites.Anim Nutr. 2024 Mar 21;17:358-372. doi: 10.1016/j.aninu.2024.01.011. eCollection 2024 Jun. Anim Nutr. 2024. PMID: 38800732 Free PMC article.
-
Dicoumarol: from chemistry to antitumor benefits.Chin Med. 2022 Dec 27;17(1):145. doi: 10.1186/s13020-022-00699-0. Chin Med. 2022. PMID: 36575479 Free PMC article. Review.
-
Natural products for infectious microbes and diseases: an overview of sources, compounds, and chemical diversities.Sci China Life Sci. 2022 Jun;65(6):1123-1145. doi: 10.1007/s11427-020-1959-5. Epub 2021 Oct 21. Sci China Life Sci. 2022. PMID: 34705221 Free PMC article. Review.
-
GlmU inhibitor from the roots of Euphorbia ebracteolata as an anti-tuberculosis agent.RSC Adv. 2022 Jun 22;12(28):18266-18273. doi: 10.1039/d2ra02044k. eCollection 2022 Jun 14. RSC Adv. 2022. PMID: 35800323 Free PMC article.
-
Biosynthesis of uridine diphosphate N-Acetylglucosamine: An underexploited pathway in the search for novel antibiotics?IUBMB Life. 2022 Dec;74(12):1232-1252. doi: 10.1002/iub.2664. Epub 2022 Jul 26. IUBMB Life. 2022. PMID: 35880704 Free PMC article. Review.
References
-
- Betts J. C., Mclaren A., Lennon M. G., Kelly F. M., Lukey P. T., Blakemore S. J., et al. (2003). Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47 2903–2913. 10.1128/aac.47.9.2903-2913.2003 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

