Engineering of a Red Fluorogenic Protein/Merocyanine Complex for Live-Cell Imaging
- PMID: 31482666
- PMCID: PMC7379159
- DOI: 10.1002/cbic.201900428
Engineering of a Red Fluorogenic Protein/Merocyanine Complex for Live-Cell Imaging
Abstract
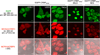
A reengineered human cellular retinol binding protein II (hCRBPII), a 15-kDa protein belonging to the intracellular lipid binding protein (iLBP) family, generates a highly fluorescent red pigment through the covalent linkage of a merocyanine aldehyde to an active site lysine residue. The complex exhibits "turn-on" fluorescence, due to a weakly fluorescent aldehyde that "lights up" with subsequent formation of a strongly fluorescent merocyanine dye within the binding pocket of the protein. Cellular penetration of merocyanine is rapid, and fluorophore maturation is nearly instantaneous. The hCRBPII/merocyanine complex displays high quantum yield, low cytotoxicity, specificity in labeling organelles, and compatibility in both cancer cell lines and yeast cells. The hCRBPII/merocyanine tag is brighter than most common red fluorescent proteins.
Keywords: hCRBPII; imaging; merocyanine; protein engineering; red fluorescence.
© 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures



References
-
- Bruchez MP, Curr. Opin. Chem. Biol. 2015, 27, 18–23; - PMC - PubMed
- Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA, Physiol. Rev. 2010, 90, 1103–1163; - PubMed
- Day RN, Davidson MW, Chem. Soc. Rev. 2009, 38, 2887–2921; - PMC - PubMed
- Enterina JR, Wu L, Campbell RE, Curr. Opin. Chem. Biol. 2015, 27, 10–17; - PubMed
- Mishin AS, Belousov VV, Solntsev KM, Lukyanov KA, Curr. Opin. Chem. Biol. 2015, 27, 1–9; - PubMed
- Newman RH, Fosbrink MD, Zhang J, Chem. Rev. 2011, 111, 3614–3666; - PMC - PubMed
- K. Nienhaus G. U.Nienhaus, Chem. Soc. Rev. 2014, 43, 1088–1106; - PubMed
- Okumoto S, Jones A, Frommer WB, Annu. Rev. Plant Biol. 2012, 63, 663–706; - PubMed
- Shcherbakova DM, Verkhusha VV, Curr. Opin. Chem. Biol. 2014, 20, 60–68. - PMC - PubMed
-
- Griffin BA, Adams SR, Tsien RY, Science 1998, 281, 269–272; - PubMed
- Hoffmann C, Gaietta G, Zurn A, Adams SR, Terrillon S, Ellisman MH, Tsien RY, Lohse MJ, Nat. Protoc. 2010, 5, 1666–1677; - PMC - PubMed
- Jing C, Cornish VW, Acc. Chem. Res. 2011, 44, 784–792; - PMC - PubMed
- Scheck RA, Schepartz A, Acc. Chem. Res. 2011, 44, 654–665. - PMC - PubMed
-
- England CG, Luo H, Cai W, Bioconjugate Chem. 2015, 26, 975–986; - PMC - PubMed
- Liu DS, Phipps WS, Loh KH, Howarth M, Ting AY, ACS Nano 2012, 6, 11080–11087; - PMC - PubMed
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohane RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV, ACS Chem. Biol. 2008, 3, 373–382; - PubMed
- Tseng J-C, Benink HA, McDougall MG, Chico-Calero I, Kung AL, Curr. Chem. Genomics 2012, 6, 48–54. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

