Association of Germline Variants in Natural Killer Cells With Tumor Immune Microenvironment Subtypes, Tumor-Infiltrating Lymphocytes, Immunotherapy Response, Clinical Outcomes, and Cancer Risk
- PMID: 31483464
- PMCID: PMC6727785
- DOI: 10.1001/jamanetworkopen.2019.9292
Association of Germline Variants in Natural Killer Cells With Tumor Immune Microenvironment Subtypes, Tumor-Infiltrating Lymphocytes, Immunotherapy Response, Clinical Outcomes, and Cancer Risk
Erratum in
-
Error in Methods.JAMA Netw Open. 2020 Apr 1;3(4):e206708. doi: 10.1001/jamanetworkopen.2020.6708. JAMA Netw Open. 2020. PMID: 32329767 Free PMC article. No abstract available.
Abstract
Importance: Only a small fraction of patients with cancer receiving immune checkpoint therapy (ICT) respond, which is associated with tumor immune microenvironment (TIME) subtypes and tumor-infiltrating lymphocytes (TILs).
Objective: To examine whether germline variants of natural killer (NK) cells, a key component of the immune system, are associated with TIME subtypes, the abundance of TILs, response to ICT, clinical outcomes, and cancer risk.
Design, setting, and participants: This genetic association study explored TIME subtypes and examined the association of the germline genomic information of patients with cancer with TIME subtypes, abundance of TILs, response to ICT, prognosis, and cancer risk. Clinical information, tumor RNA sequencing, and whole-exome sequencing (WES) data of paired normal samples of patients with 13 common cancers (n = 5883) were obtained from the Cancer Genome Atlas. The WES data of individuals with no cancer (n = 4500) were obtained from the database of Genotypes and Phenotypes. Data collection and analysis took place in March 2017.
Main outcomes and measures: Associations between the number of germline defective genes in NK cells and survival time and the abundance of TILs.
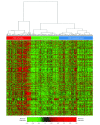
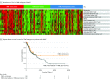
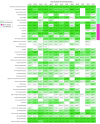
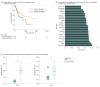
Results: Based on tumor RNA sequencing data, tumors were stratified into TIME-rich, TIME-intermediate, and TIME-poor subtypes. Tumors of TIME-rich subtype had more TILs (TIL-NK cells in TIME-rich head and neck squamous cell carcinoma [HNSC] tumors: t = 4.85; 95% CI of the difference, 0.01-0.03; P = 2.19 × 10-6) compared with TIME-intermediate HNSC tumors (t = 3.70; 95% CI of the difference, 0.01-0.03; P < .001), better prognosis (patients with HNSC: hazard ratio, 0.65; 95% CI, 0.41-1.02; P = .054) compared with TIME-intermediate and TIME-poor subtypes, and better ICT response (patients with melanoma: odds ratio [OR], 4.45; 95% CI, 0.99-27.08; P = .04). Patients with TIME-rich tumors had significantly fewer inherited defective genes in NK cells than patients with TIME-intermediate and TIME-poor tumors (patients with HNSC: OR, 0.49; 95% CI, 0.26-1.07; P = .005). Similarly, patients with cancer had significantly more inherited defective genes in NK cells than individuals with no cancer (patients with HNSC: OR, 19.09; 95% CI, 4.30-315.96; P = 6.21 × 10-4). Among 11 of 13 common cancers, the number of heritable defective genes in NK cells was significantly negatively associated with survival (patients with HNSC: hazard ratio, 1.77; 95% CI, 1.18-2.66; P = .005), abundance of TILs (patients with HNSC: R = -0.25; 95% CI, -0.65-2.17; P = 0.02), and response to ICT (patients with melanoma: OR, 4.45; 95% CI, 0.99-27.08; P = .04).
Conclusions and relevance: These results suggest that individuals who have more inherited defective genes in NK cells had a higher risk of developing cancer and that these inherited defects were associated with TIME subtypes, recruitment of TILs, ICT response, and clinical outcomes. The findings have implications for identifying individuals at risk for developing cancer of many types based on germline variants of NK cells and for improving existing ICT and chimeric antigen receptor-T cell therapy by adoptive transfer of healthy NK cells to patients with TIME-intermediate and TIME-poor tumors.
Conflict of interest statement
Figures

Similar articles
-
Characterization of tumor-infiltrating lymphocytes and their spatial distribution in triple-negative breast cancer.Breast Cancer Res. 2024 Dec 6;26(1):180. doi: 10.1186/s13058-024-01932-4. Breast Cancer Res. 2024. PMID: 39643914 Free PMC article.
-
Connecting the Dots: Therapy-Induced Senescence and a Tumor-Suppressive Immune Microenvironment.J Natl Cancer Inst. 2015 Dec 30;108(6):djv406. doi: 10.1093/jnci/djv406. Print 2016 Jun. J Natl Cancer Inst. 2015. PMID: 26719346 Free PMC article.
-
Reduction of immunosuppressive tumor microenvironment in cholangiocarcinoma by ex vivo targeting immune checkpoint molecules.J Hepatol. 2019 Oct;71(4):753-762. doi: 10.1016/j.jhep.2019.05.026. Epub 2019 Jun 11. J Hepatol. 2019. PMID: 31195061
-
Prognostic and therapeutic role of tumor-infiltrating lymphocyte subtypes in breast cancer.Cancer Metastasis Rev. 2021 Jun;40(2):519-536. doi: 10.1007/s10555-021-09968-0. Epub 2021 May 7. Cancer Metastasis Rev. 2021. PMID: 33963482 Free PMC article. Review.
-
Variation of tumor-infiltrating lymphocytes in human cancers: controversy on clinical significance.Immunotherapy. 2011 Oct;3(10):1235-51. doi: 10.2217/imt.11.106. Immunotherapy. 2011. PMID: 21995574 Review.
Cited by
-
The Expression Quantitative Trait Loci in Immune Response Genes Impact the Characteristics and Survival of Colorectal Cancer.Diagnostics (Basel). 2022 Jan 26;12(2):315. doi: 10.3390/diagnostics12020315. Diagnostics (Basel). 2022. PMID: 35204406 Free PMC article.
-
Error in Methods.JAMA Netw Open. 2020 Apr 1;3(4):e206708. doi: 10.1001/jamanetworkopen.2020.6708. JAMA Netw Open. 2020. PMID: 32329767 Free PMC article. No abstract available.
-
Abnormal Expression of c-Myc Oncogene in NK Cells in Patients with Cancer.Int J Mol Sci. 2019 Feb 11;20(3):756. doi: 10.3390/ijms20030756. Int J Mol Sci. 2019. PMID: 30754645 Free PMC article.
-
CD2 Is a Novel Immune-Related Prognostic Biomarker of Invasive Breast Carcinoma That Modulates the Tumor Microenvironment.Front Immunol. 2021 Apr 23;12:664845. doi: 10.3389/fimmu.2021.664845. eCollection 2021. Front Immunol. 2021. PMID: 33968066 Free PMC article.
-
Natural Killer Cell Therapy: A New Treatment Paradigm for Solid Tumors.Cancers (Basel). 2019 Oct 11;11(10):1534. doi: 10.3390/cancers11101534. Cancers (Basel). 2019. PMID: 31614472 Free PMC article. Review.

