Ultrafast magnetic resonance spectroscopic imaging using SPICE with learned subspaces
- PMID: 31483526
- PMCID: PMC6824949
- DOI: 10.1002/mrm.27980
Ultrafast magnetic resonance spectroscopic imaging using SPICE with learned subspaces
Abstract
Purpose: To develop a subspace learning method for the recently proposed subspace-based MRSI approach known as SPICE, and achieve ultrafast 1 H-MRSI of the brain.
Theory and methods: A novel strategy is formulated to learn a low-dimensional subspace representation of MR spectra from specially acquired training data and use the learned subspace for general MRSI experiments. Specifically, the subspace learning problem is formulated as learning "empirical" distributions of molecule-specific spectral parameters (e.g., concentrations, lineshapes, and frequency shifts) by integrating physics-based model and the training data. The learned spectral parameters and quantum mechanical simulation basis can then be combined to construct acquisition-specific subspace for spatiospectral encoding and processing. High-resolution MRSI acquisitions combining ultrashort-TE/short-TR excitation, sparse sampling, and the elimination of water suppression have been performed to evaluate the feasibility of the proposed method.
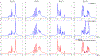
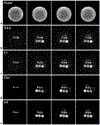
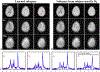
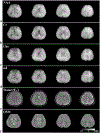
Results: The accuracy of the learned subspace and the capability of the proposed method in producing high-resolution 3D 1 H metabolite maps and high-quality spatially resolved spectra (with a nominal resolution of ∼2.4 × 2.4 × 3 mm3 in 5 minutes) were demonstrated using phantom and in vivo studies. By eliminating water suppression, we are also able to extract valuable information from the water signals for data processing ( map, frequency drift, and coil sensitivity) as well as for mapping tissue susceptibility and relaxation parameters.
Conclusions: The proposed method enables ultrafast 1 H-MRSI of the brain using a learned subspace, eliminating the need of acquiring subject-dependent navigator data (known as ) in the original SPICE technique. It represents a new way to perform MRSI experiments and an important step toward practical applications of high-resolution MRSI.
Keywords: MR spectroscopic imaging; no water suppression; rapid spatiospectral encoding; subspace learning; union-of-subspaces model.
© 2019 International Society for Magnetic Resonance in Medicine.
Figures
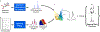

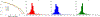
References
-
- Lauterbur PC, Kramer DM, House WV, Chen CN, Zeugmatographic high resolution nuclear magnetic resonance spectroscopy: Images of chemical inhomogeneity within macroscopic objects. J Amer Chem Soc 1975;97:6866 – 6868.
-
- Maudsley AA, Hilal SK, Perman WH, Simon HE, Spatially resolved high resolution spectroscopy by “four-dimensional” NMR. J Magn Reson 1983;51:147 – 152.
-
- Luyten PR, Marien AJ, Heindel W, van Gerwen PH, Herholz K, den Hollander JA, Friedmann G, Heiss WD, Metabolic imaging of patients with intracranial tumors: H-1 MR spectroscopic imaging and PET. Radiology 1990;176:791 – 799. - PubMed
-
- de Graaf RA, In Vivo NMR Spectroscopy: Principles and Techniques. Hoboken, NJ: John Wiley and Sons, 2007.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

