DdlR, an essential transcriptional regulator of peptidoglycan biosynthesis in Clostridioides difficile
- PMID: 31483905
- PMCID: PMC6842417
- DOI: 10.1111/mmi.14371
DdlR, an essential transcriptional regulator of peptidoglycan biosynthesis in Clostridioides difficile
Abstract
D-Ala-D-Ala ligase, encoded by ddl genes, is responsible for the synthesis of a dipeptide, D-Ala-D-Ala, an essential precursor of bacterial peptidoglycan. In Clostridioides difficile, the single ddl gene is located upstream of the ddlR gene, which encodes a putative transcriptional regulator. Using mutational and transcriptional analysis and DNA-binding assays, DdlR was found to be a direct activator of the ddl ddlR operon. DdlR is a member of the MocR/GabR-type proteins that have aminotransferase-like, pyridoxal 5'-phosphate-binding domains. A DdlR mutation that prevented covalent binding of pyridoxal 5'-phosphate abolished the ability of DdlR to activate transcription. Addition of D-Ala-D-Ala to the medium inactivated DdlR, reducing dipeptide biosynthesis. In contrast, D-Ala-D-Ala limitation caused a dramatic increase in expression from the ddl promoter. Though uncommon for transcription regulators, C. difficile DdlR is essential, as the ddlR null mutant cells could not grow even in complex laboratory media in the absence of D-Ala-D-Ala. A dyad symmetry sequence, which is located immediately upstream of the -35 region of the ddl promoter, serves as an important element of the DdlR-binding site. This sequence is conserved upstream of putative DdlR targets in other bacteria of classes Clostridia and Bacilli, indicating a similar mode of regulation of these genes.
© 2019 John Wiley & Sons Ltd.
Figures
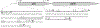
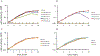



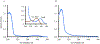

References
-
- Bailey TL and Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol, 2, 28–36. - PubMed
-
- Barreteau H, Kovac A, Boniface A, Sova M, Gobec S and Blanot D (2008) Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev, 32, 168–207. - PubMed
-
- Belitsky BR (2004) Bacillus subtilis GabR, a protein with DNA-binding and aminotransferase domains, is a PLP-dependent transcriptional regulator. J. Mol. Biol, 340, 655–664. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

