Cross Talk between SigB and PrfA in Listeria monocytogenes Facilitates Transitions between Extra- and Intracellular Environments
- PMID: 31484692
- PMCID: PMC6759667
- DOI: 10.1128/MMBR.00034-19
Cross Talk between SigB and PrfA in Listeria monocytogenes Facilitates Transitions between Extra- and Intracellular Environments
Abstract
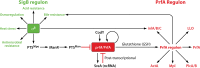
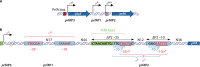
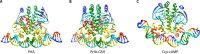
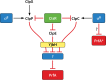
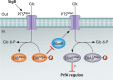
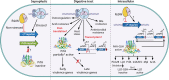
The foodborne pathogen Listeria monocytogenes can modulate its transcriptome and proteome to ensure its survival during transmission through vastly differing environmental conditions. While L. monocytogenes utilizes a large array of regulators to achieve survival and growth in different intra- and extrahost environments, the alternative sigma factor σB and the transcriptional activator of virulence genes protein PrfA are two key transcriptional regulators essential for responding to environmental stress conditions and for host infection. Importantly, emerging evidence suggests that the shift from extrahost environments to the host gastrointestinal tract and, subsequently, to intracellular environments requires regulatory interplay between σB and PrfA at transcriptional, posttranscriptional, and protein activity levels. Here, we review the current evidence for cross talk and interplay between σB and PrfA and their respective regulons and highlight the plasticity of σB and PrfA cross talk and the role of this cross talk in facilitating successful transition of L. monocytogenes from diverse extrahost to diverse extra- and intracellular host environments.
Keywords: Listeria monocytogenes; PrfA; gene regulation; general stress response; sigma B; virulence.
Copyright © 2019 American Society for Microbiology.
Figures
References
-
- Rouquette C, Berche P. 1996. The pathogenesis of infection by Listeria monocytogenes. Microbiologia 12:245–258. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

