The SH3 domains of the protein kinases ITK and LCK compete for adjacent sites on T cell-specific adapter protein
- PMID: 31484725
- PMCID: PMC6802506
- DOI: 10.1074/jbc.RA119.008318
The SH3 domains of the protein kinases ITK and LCK compete for adjacent sites on T cell-specific adapter protein
Abstract
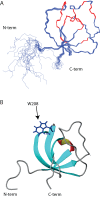
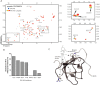
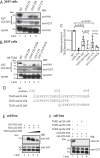
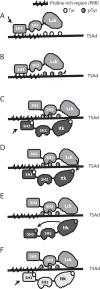
T-cell activation requires stimulation of specific intracellular signaling pathways in which protein-tyrosine kinases, phosphatases, and adapter proteins interact to transmit signals from the T-cell receptor to the nucleus. Interactions of LCK proto-oncogene, SRC family tyrosine kinase (LCK), and the IL-2-inducible T cell kinase (ITK) with the T cell-specific adapter protein (TSAD) promotes LCK-mediated phosphorylation and thereby ITK activation. Both ITK and LCK interact with TSAD's proline-rich region (PRR) through their Src homology 3 (SH3) domains. Whereas LCK may also interact with TSAD through its SH2 domain, ITK interacts with TSAD only through its SH3 domain. To begin to understand on a molecular level how the LCK SH3 and ITK SH3 domains interact with TSAD in human HEK293T cells, here we combined biochemical analyses with NMR spectroscopy. We found that the ITK and LCK SH3 domains potentially have adjacent and overlapping binding sites within the TSAD PRR amino acids (aa) 239-274. Pulldown experiments and NMR spectroscopy revealed that both domains may bind to TSAD aa 239-256 and aa 257-274. Co-immunoprecipitation experiments further revealed that both domains may also bind simultaneously to TSAD aa 242-268. Accordingly, NMR spectroscopy indicated that the SH3 domains may compete for these two adjacent binding sites. We propose that once the associations of ITK and LCK with TSAD promote the ITK and LCK interaction, the interactions among TSAD, ITK, and LCK are dynamically altered by ITK phosphorylation status.
Keywords: IL-2-inducible T cell kinase (ITK); LCK proto-oncogene SRC family tyrosine kinase; NMR; SH2D2A; Src homology 3 domain (SH3 domain); T cell-specific adapter protein (TSAD); T-cell; adaptor protein; cell signaling; immunity; nuclear magnetic resonance; protein kinase; protein phosphorylation; protein structure; protein-protein interaction; tyrosine-protein kinase.
© 2019 Andersen et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- Spurkland A., Brinchmann J. E., Markussen G., Pedeutour F., Munthe E., Lea T., Vartdal F., and Aasheim H. C. (1998) Molecular cloning of a T cell-specific adapter protein (TSAD) containing an Src homology (SH) 2 domain and putative SH3 and phosphotyrosine binding sites. J. Biol. Chem. 273, 4539–4546 10.1074/jbc.273.8.4539 - DOI - PubMed
-
- Granum S., Sundvold-Gjerstad V., Dai K. Z., Kolltveit K. M., Hildebrand K., Huitfeldt H. S., Lea T., and Spurkland A. (2006) Structure function analysis of SH2D2A isoforms expressed in T cells reveals a crucial role for the proline rich region encoded by SH2D2A exon 7. BMC. Immunol. 7, 15 10.1186/1471-2172-7-15 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

