Examining the Evidence for an Adult Healthy Middle Ear Microbiome
- PMID: 31484741
- PMCID: PMC6731531
- DOI: 10.1128/mSphere.00456-19
Examining the Evidence for an Adult Healthy Middle Ear Microbiome
Abstract
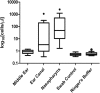
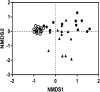
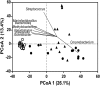
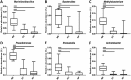
Otitis media (OM) is a cluster of diseases of the middle ear that commonly result from bacterial infection. OM subtypes in which the tympanic membrane is intact (acute otitis media and otitis media with effusion) are presumed to result from pathogen translocation through the eustachian tube. Recent molecular-based studies have suggested that a diverse middle ear microbiome exists in the absence of disease. These have been largely unsupported by culture and feature species that commonly contaminate low-biomass sequencing data. Combining culture-based and molecular techniques, we undertook a detailed investigation of the evidence for bacterial colonization of the healthy middle ear. Middle ear (ME), nasopharynx (NP), and external ear canal (EC) swabs were collected from a total of 25 adult patients undergoing cochlear implant, stapedotomy, or translabyrinthine vestibular schwannoma resection. Diagnostic culture, microscopy, quantitative PCR, and 16S rRNA gene amplicon sequencing were used to assess sample bacterial content. EC and NP microbiota were consistent with previous reports. In contrast, bacterial levels in ME samples were not significantly above those in unused control swabs. Commonly detected taxa were among recognized sequencing contaminants (Methylobacterium, Pseudomonas, and Acinetobacter). Linear regression of dominant ME taxa confirmed a negative relationship between relative abundance and bacterial load, consistent with contamination. No bacteria were detected by microscopy or diagnostic culture in any middle ear sample. Our findings cast substantial doubt on previous reports identifying a healthy middle ear microbiome using 16S amplicon sequencing.IMPORTANCE Recent molecular-based studies have suggested that a diverse middle ear microbiome in adults and children can exist in the absence of disease. These studies have been largely unsupported by culture and feature species that commonly contaminate low-biomass sequencing data. While 16S rRNA gene amplicon sequencing has proven to be a highly informative technique in many clinical contexts, it is susceptible to spurious signal arising from sequencing reagent contaminants where sample biomass is low. Combining culture-based and molecular techniques, we undertook a detailed investigation of the evidence for bacterial colonization of the healthy middle ear. In finding no evidence of viable bacterial cells in middle ear samples, our study further underlines the importance of careful consideration of amplicon sequence data derived from very-low-biomass contexts and the value of analytical approaches that combine culture and molecular techniques.
Keywords: microbiome; middle ear; otitis media.
Copyright © 2019 Jervis-Bardy et al.
Figures
References
-
- Vergison A, Dagan R, Arguedas A, Bonhoeffer J, Cohen R, Dhooge I, Hoberman A, Liese J, Marchisio P, Palmu AA, Ray GT, Sanders EA, Simoes EA, Uhari M, van Eldere J, Pelton SI. 2010. Otitis media and its consequences: beyond the earache. Lancet Infect Dis 10:195–203. doi: 10.1016/S1473-3099(10)70012-8. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

