Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells
- PMID: 31484774
- PMCID: PMC6765243
- DOI: 10.1073/pnas.1908626116
Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells
Abstract
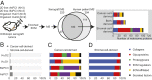
Pancreatic ductal adenocarcinoma (PDAC) has prominent extracellular matrix (ECM) that compromises treatments yet cannot be nonselectively disrupted without adverse consequences. ECM of PDAC, despite the recognition of its importance, has not been comprehensively studied in patients. In this study, we used quantitative mass spectrometry (MS)-based proteomics to characterize ECM proteins in normal pancreas and pancreatic intraepithelial neoplasia (PanIN)- and PDAC-bearing pancreas from both human patients and mouse genetic models, as well as chronic pancreatitis patient samples. We describe detailed changes in both abundance and complexity of matrisome proteins in the course of PDAC progression. We reveal an early up-regulated group of matrisome proteins in PanIN, which are further up-regulated in PDAC, and we uncover notable similarities in matrix changes between pancreatitis and PDAC. We further assigned cellular origins to matrisome proteins by performing MS on multiple lines of human-to-mouse xenograft tumors. We found that, although stromal cells produce over 90% of the ECM mass, elevated levels of ECM proteins derived from the tumor cells, but not those produced exclusively by stromal cells, tend to correlate with poor patient survival. Furthermore, distinct pathways were implicated in regulating expression of matrisome proteins in cancer cells and stromal cells. We suggest that, rather than global suppression of ECM production, more precise ECM manipulations, such as targeting tumor-promoting ECM proteins and their regulators in cancer cells, could be more effective therapeutically.
Keywords: ECM; PDAC; PanIN; pancreatitis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 (2018). - PubMed
-
- Swartz M. A., Lund A. W., Lymphatic and interstitial flow in the tumour microenvironment: Linking mechanobiology with immunity. Nat. Rev. Cancer 12, 210–219 (2012). - PubMed
-
- Hartmann N., et al. , Prevailing role of contact guidance in intrastromal T-cell trapping in human pancreatic cancer. Clin. Cancer. Res. 20, 3422–3433 (2014). - PubMed
-
- Yu M., Tannock I. F., Targeting tumor architecture to favor drug penetration: A new weapon to combat chemoresistance in pancreatic cancer? Cancer Cell 21, 327–329 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA190092/CA/NCI NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- U10 CA180944/CA/NCI NIH HHS/United States
- P30 CA045508/CA/NCI NIH HHS/United States
- U01 CA214125/CA/NCI NIH HHS/United States
- P20 CA192996/CA/NCI NIH HHS/United States
- U24 CA160034/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U24 CA210986/CA/NCI NIH HHS/United States
- P20 CA192994/CA/NCI NIH HHS/United States
- U24 CA126476/CA/NCI NIH HHS/United States
- U01 CA210240/CA/NCI NIH HHS/United States
- U24 CA210979/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

