A Prospective Analysis of Genetic Variants Associated with Human Lifespan
- PMID: 31484785
- PMCID: PMC6723124
- DOI: 10.1534/g3.119.400448
A Prospective Analysis of Genetic Variants Associated with Human Lifespan
Abstract
We present a massive investigation into the genetic basis of human lifespan. Beginning with a genome-wide association (GWA) study using a de-identified snapshot of the unique AncestryDNA database - more than 300,000 genotyped individuals linked to pedigrees of over 400,000,000 people - we mapped six genome-wide significant loci associated with parental lifespan. We compared these results to a GWA analysis of the traditional lifespan proxy trait, age, and found only one locus, APOE, to be associated with both age and lifespan. By combining the AncestryDNA results with those of an independent UK Biobank dataset, we conducted a meta-analysis of more than 650,000 individuals and identified fifteen parental lifespan-associated loci. Beyond just those significant loci, our genome-wide set of polymorphisms accounts for up to 8% of the variance in human lifespan; this value represents a large fraction of the heritability estimated from phenotypic correlations between relatives.
Keywords: GWAS; human; lifespan.
Copyright © 2019 Wright et al.
Figures
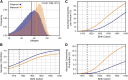


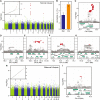

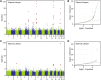
References
-
- Ball C., Barber M., Byrnes J., Callaway J., Chahine K. et al. , 2013. Ethnicity Estimate White Paper. https://www.ancestry.com/dna/resource/whitePaper/AncestryDNA-Ethnicity-W....
-
- Ball C., Barber M., Byrnes J., Carbonetto P., Chahine K. et al. , 2016. Ancestry DNA Matching White Paper. https://www.ancestry.com/corporate/sites/default/files/AncestryDNA-Match....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
