CD36 inhibits β-catenin/c-myc-mediated glycolysis through ubiquitination of GPC4 to repress colorectal tumorigenesis
- PMID: 31484922
- PMCID: PMC6726635
- DOI: 10.1038/s41467-019-11662-3
CD36 inhibits β-catenin/c-myc-mediated glycolysis through ubiquitination of GPC4 to repress colorectal tumorigenesis
Abstract
The diverse expression pattern of CD36 reflects its multiple cellular functions. However, the roles of CD36 in colorectal cancer (CRC) remain unknown. Here, we discover that CD36 expression is progressively decreased from adenomas to carcinomas. CD36 loss predicts poor survival of CRC patients. In CRC cells, CD36 acts as a tumor suppressor and inhibits aerobic glycolysis in vitro and in vivo. Mechanically, CD36-Glypcian 4 (GPC4) interaction could promote the proteasome-dependent ubiquitination of GPC4, followed by inhibition of β-catenin/c-myc signaling and suppression of downstream glycolytic target genes GLUT1, HK2, PKM2 and LDHA. Moreover, disruption of CD36 in inflammation-induced CRC model as well as ApcMin/+ mice model significantly increased colorectal tumorigenesis. Our results reveal a CD36-GPC4-β-catenin-c-myc signaling axis that regulates glycolysis in CRC development and may provide an intervention strategy for CRC prevention.
Conflict of interest statement
The authors declare no competing interests.
Figures


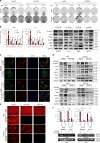
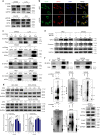
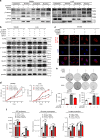
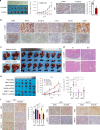

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

