Photo-editable macromolecular information
- PMID: 31484927
- PMCID: PMC6726599
- DOI: 10.1038/s41467-019-11566-2
Photo-editable macromolecular information
Abstract
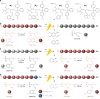
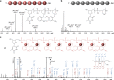
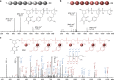
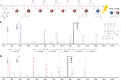
Light-induced alteration of macromolecular information plays a central role in biology and is known to influence health, aging and Darwinian evolution. Here, we report that light can also trigger sequence variations in abiotic information-containing polymers. Sequence-coded poly(phosphodiester)s were synthesized using four phosphoramidite monomers containing either photo-sensitive or photo-inert substituents. These monomers allow different sequence manipulations. For instance, using two light-cleavable monomers containing o-nitrobenzyl ether and o-nitroveratryl ether motifs, photo-erasable digital polymers were prepared. These polymers can be decoded by tandem mass spectrometry but become unreadable after UVA exposure. The opposite behavior, i.e. photo-revealable sequences, was obtained with polymers made of two isobaric monomers containing light-cleavable o-nitrobenzyl ether and light-inert p-nitrobenzyl ether substituents. Furthermore, when the latter two monomers were used in conjunction with a third monomer bearing a light-inert OH group, site-directed photo-mutations were induced in synthetic polymers. This was used herein to change the meaning of binary sequences.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Lutz J-F. Coding macromolecules: inputting information in polymers using monomer-based alphabets. Macromolecules. 2015;48:4759–4767. doi: 10.1021/acs.macromol.5b00890. - DOI
-
- Rutten, M. G. T. A., Vaandrager, F. W., Elemans, J. A. A. W. & Nolte, R. J. M. Encoding information into polymers. Nat. Rev. Chem. 2, 365–381 (2018).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

