Activation of PKA via asymmetric allosteric coupling of structurally conserved cyclic nucleotide binding domains
- PMID: 31484930
- PMCID: PMC6726620
- DOI: 10.1038/s41467-019-11930-2
Activation of PKA via asymmetric allosteric coupling of structurally conserved cyclic nucleotide binding domains
Abstract
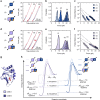
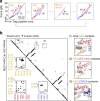
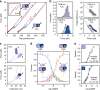
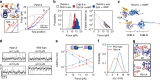
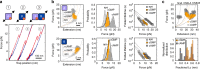
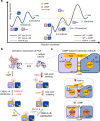
Cyclic nucleotide-binding (CNB) domains allosterically regulate the activity of proteins with diverse functions, but the mechanisms that enable the cyclic nucleotide-binding signal to regulate distant domains are not well understood. Here we use optical tweezers and molecular dynamics to dissect changes in folding energy landscape associated with cAMP-binding signals transduced between the two CNB domains of protein kinase A (PKA). We find that the response of the energy landscape upon cAMP binding is domain specific, resulting in unique but mutually coordinated tasks: one CNB domain initiates cAMP binding and cooperativity, whereas the other triggers inter-domain interactions that promote the active conformation. Inter-domain interactions occur in a stepwise manner, beginning in intermediate-liganded states between apo and cAMP-bound domains. Moreover, we identify a cAMP-responsive switch, the N3A motif, whose conformation and stability depend on cAMP occupancy. This switch serves as a signaling hub, amplifying cAMP-binding signals during PKA activation.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Mayer BJ. SH3 domains: complexity in moderation. J. Cell. Sci. 2001;114:1253–1263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

