Agent-based modeling and bifurcation analysis reveal mechanisms of macrophage polarization and phenotype pattern distribution
- PMID: 31484958
- PMCID: PMC6726649
- DOI: 10.1038/s41598-019-48865-z
Agent-based modeling and bifurcation analysis reveal mechanisms of macrophage polarization and phenotype pattern distribution
Abstract
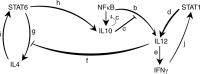
Macrophages play a key role in tissue regeneration by polarizing to different destinies and generating various phenotypes. Recognizing the underlying mechanisms is critical in designing therapeutic procedures targeting macrophage fate determination. Here, to investigate the macrophage polarization, a nonlinear mathematical model is proposed in which the effect of IL4, IFNγ and LPS, as external stimuli, on STAT1, STAT6, and NFκB is studied using bifurcation analysis. The existence of saddle-node bifurcations in these internal key regulators allows different combinations of steady state levels which are attributable to different fates. Therefore, we propose dynamic bifurcation as a crucial built-in mechanism of macrophage polarization. Next, in order to investigate the polarization of a population of macrophages, bifurcation analysis is employed aligned with agent-based approach and a two-layer model is proposed in which the information from single cells is exploited to model the behavior in tissue level. Also, in this model, a partial differential equation describes the diffusion of secreted cytokines in the medium. Finally, the model was validated against a set of experimental data. Taken together, we have here developed a cell and tissue level model of macrophage polarization behavior which can be used for designing therapeutic interventions.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Osteoclast stimulatory transmembrane protein induces a phenotypic switch in macrophage polarization suppressing an M1 pro-inflammatory state.Acta Biochim Biophys Sin (Shanghai). 2017 Oct 1;49(10):935-944. doi: 10.1093/abbs/gmx092. Acta Biochim Biophys Sin (Shanghai). 2017. PMID: 28981605
-
Pentamethoxyflavanone regulates macrophage polarization and ameliorates sepsis in mice.Biochem Pharmacol. 2014 May 1;89(1):109-18. doi: 10.1016/j.bcp.2014.02.016. Epub 2014 Mar 4. Biochem Pharmacol. 2014. PMID: 24607272
-
Transcription factors STAT6 and KLF4 implement macrophage polarization via the dual catalytic powers of MCPIP.J Immunol. 2015 Jun 15;194(12):6011-23. doi: 10.4049/jimmunol.1402797. Epub 2015 May 1. J Immunol. 2015. PMID: 25934862 Free PMC article.
-
Physalin D regulates macrophage M1/M2 polarization via the STAT1/6 pathway.J Cell Physiol. 2019 Jun;234(6):8788-8796. doi: 10.1002/jcp.27537. Epub 2018 Oct 14. J Cell Physiol. 2019. PMID: 30317606
-
Transcriptional regulation of macrophage polarization: enabling diversity with identity.Nat Rev Immunol. 2011 Oct 25;11(11):750-61. doi: 10.1038/nri3088. Nat Rev Immunol. 2011. PMID: 22025054 Review.
Cited by
-
An agent-based model of monocyte differentiation into tumour-associated macrophages in chronic lymphocytic leukemia.iScience. 2023 May 19;26(6):106897. doi: 10.1016/j.isci.2023.106897. eCollection 2023 Jun 16. iScience. 2023. PMID: 37332613 Free PMC article.
-
In silico assessment of CAR macrophages activity against SARS-CoV-2 infection.Heliyon. 2024 Oct 23;10(21):e39689. doi: 10.1016/j.heliyon.2024.e39689. eCollection 2024 Nov 15. Heliyon. 2024. PMID: 39524874 Free PMC article.
-
The double-edged sword role of fibroblasts in the interaction with cancer cells; an agent-based modeling approach.PLoS One. 2020 May 8;15(5):e0232965. doi: 10.1371/journal.pone.0232965. eCollection 2020. PLoS One. 2020. PMID: 32384110 Free PMC article.
-
Agent-based modeling of macrophage-fibroblast interactions in the immune response to biomaterials.PLoS One. 2025 Aug 19;20(8):e0329186. doi: 10.1371/journal.pone.0329186. eCollection 2025. PLoS One. 2025. PMID: 40828805 Free PMC article.
-
Uncovering bifurcation behaviors of biochemical reaction systems from network topology.Sci Rep. 2025 Jul 29;15(1):27596. doi: 10.1038/s41598-025-10688-6. Sci Rep. 2025. PMID: 40730575 Free PMC article.
References
-
- Fraternale A, Brundu S, Magnani M. Polarization and repolarization of macrophages. J Clin Cell Immunol. 2015;6:2–12.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

