The poly-SUMO2/3 protease SENP6 enables assembly of the constitutive centromere-associated network by group deSUMOylation
- PMID: 31485003
- PMCID: PMC6726658
- DOI: 10.1038/s41467-019-11773-x
The poly-SUMO2/3 protease SENP6 enables assembly of the constitutive centromere-associated network by group deSUMOylation
Abstract
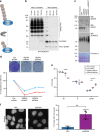
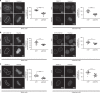
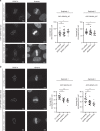
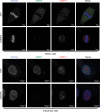
In contrast to our extensive knowledge on ubiquitin polymer signaling, we are severely limited in our understanding of poly-SUMO signaling. We set out to identify substrates conjugated to SUMO polymers, using knockdown of the poly-SUMO2/3 protease SENP6. We identify over 180 SENP6 regulated proteins that represent highly interconnected functional groups of proteins including the constitutive centromere-associated network (CCAN), the CENP-A loading factors Mis18BP1 and Mis18A and DNA damage response factors. Our results indicate a striking protein group de-modification by SENP6. SENP6 deficient cells are severely compromised for proliferation, accumulate in G2/M and frequently form micronuclei. Accumulation of CENP-T, CENP-W and CENP-A to centromeres is impaired in the absence of SENP6. Surprisingly, the increase of SUMO chains does not lead to ubiquitin-dependent proteasomal degradation of the CCAN subunits. Our results indicate that SUMO polymers can act in a proteolysis-independent manner and consequently, have a more diverse signaling function than previously expected.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Genetic screening identifies a SUMO protease dynamically maintaining centromeric chromatin.Nat Commun. 2020 Jan 24;11(1):501. doi: 10.1038/s41467-019-14276-x. Nat Commun. 2020. PMID: 31980633 Free PMC article.
-
SENP6-mediated M18BP1 deSUMOylation regulates CENP-A centromeric localization.Cell Res. 2019 Mar;29(3):254-257. doi: 10.1038/s41422-018-0139-y. Epub 2019 Jan 10. Cell Res. 2019. PMID: 30631152 Free PMC article. No abstract available.
-
Swapping small ubiquitin-like modifier (SUMO) isoform specificity of SUMO proteases SENP6 and SENP7.J Biol Chem. 2011 Oct 14;286(41):36142-36151. doi: 10.1074/jbc.M111.268847. Epub 2011 Aug 30. J Biol Chem. 2011. PMID: 21878624 Free PMC article.
-
The fate of metaphase kinetochores is weighed in the balance of SUMOylation during S phase.Cell Cycle. 2010 Aug 15;9(16):3194-201. doi: 10.4161/cc.9.16.12619. Epub 2010 Aug 9. Cell Cycle. 2010. PMID: 20724819 Free PMC article. Review.
-
A Chain of Events: Regulating Target Proteins by SUMO Polymers.Trends Biochem Sci. 2021 Feb;46(2):113-123. doi: 10.1016/j.tibs.2020.09.002. Epub 2020 Sep 29. Trends Biochem Sci. 2021. PMID: 33008689 Review.
Cited by
-
SUMOylation of PUM2 promotes the vasculogenic mimicry of glioma cells via regulating CEBPD.Clin Transl Med. 2020 Sep;10(5):e168. doi: 10.1002/ctm2.168. Clin Transl Med. 2020. PMID: 32997416 Free PMC article.
-
Gene replacement strategies validate the use of functional tags on centromeric chromatin and invalidate an essential role for CENP-AK124ub.Cell Rep. 2021 Nov 2;37(5):109924. doi: 10.1016/j.celrep.2021.109924. Cell Rep. 2021. PMID: 34731637 Free PMC article.
-
SUMO-mediated regulation of H3K4me3 reader SET-26 controls germline development in C. elegans.PLoS Biol. 2025 Jan 6;23(1):e3002980. doi: 10.1371/journal.pbio.3002980. eCollection 2025 Jan. PLoS Biol. 2025. PMID: 39761316 Free PMC article.
-
Poly-SUMO-2/3 chain modification of Nuf2 facilitates CENP-E kinetochore localization and chromosome congression during mitosis.Cell Cycle. 2021 May;20(9):855-873. doi: 10.1080/15384101.2021.1907509. Epub 2021 Apr 28. Cell Cycle. 2021. PMID: 33910471 Free PMC article.
-
In-Plate Chemical Synthesis of Isopeptide-Linked SUMOylated Peptide Fluorescence Polarization Reagents for High-Throughput Screening of SENP Preferences.Chembiochem. 2023 Jan 17;24(2):e202200601. doi: 10.1002/cbic.202200601. Epub 2022 Dec 7. Chembiochem. 2023. PMID: 36377600 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

