Analysis of clinical significance and prospective molecular mechanism of main elements of the JAK/STAT pathway in hepatocellular carcinoma
- PMID: 31485610
- PMCID: PMC6741847
- DOI: 10.3892/ijo.2019.4862
Analysis of clinical significance and prospective molecular mechanism of main elements of the JAK/STAT pathway in hepatocellular carcinoma
Abstract
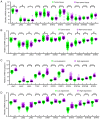
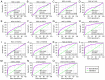
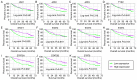
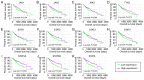
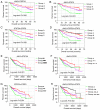
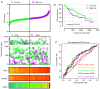
Hepatocellular carcinoma (HCC) is one the most common malignancies and has poor prognosis in patients. The aim of the present study is to explore the clinical significance of the main genes involved in the Janus kinase (JAK)‑signal transducer and activator of transcription (STAT) pathway in HCC. GSE14520, a training cohort containing 212 hepatitis B virus‑infected HCC patients from the Gene Expression Omnibus database, and data from The Cancer Genome Atlas as a validation cohort containing 370 HCC patients, were used to analyze the diagnostic and prognostic significance for HCC. Joint‑effect analyses were performed to determine diagnostic and prognostic significance. Nomograms and risk score models were constructed to predict HCC prognosis using the two cohorts. Additionally, molecular mechanism analysis was performed for the two cohorts. Prognosis‑associated genes in the two cohorts were further validated for differential expression using reverse transcription‑quantitative polymerase chain reaction of 21 pairs of hepatitis B virus‑infected HCC samples. JAK2, TYK2, STAT3, STAT4 and STAT5B had diagnostic significance in the two cohorts (all area under curves >0.5; P≤0.05). In addition, JAK2, STAT5A, STAT6 exhibited prognostic significance in both cohorts (all adjusted P≤0.05). Furthermore, joint‑effect analysis had advantages over using one gene alone. Molecular mechanism analyses confirmed that STAT6 was enriched in pathways and terms associated with the cell cycle, cell division and lipid metabolism. Nomograms and risk score models had advantages for HCC prognosis prediction. When validated in 21 pairs of HCC and non‑tumor tissue, STAT6 was differentially expressed, whereas JAK2 was not differentially expressed. In conclusion, JAK2, STAT5A and STAT6 may be potential prognostic biomarkers for HCC. JAK2, TYK2, STAT3, STAT4 and STAT5B may be potential diagnostic biomarkers for HCC. STAT6 has a role in HCC that may be mediated via effects on the cell cycle, cell division and lipid metabolism.
Figures




Similar articles
-
JAK/STAT/SOCS-signaling pathway and colon and rectal cancer.Mol Carcinog. 2013 Feb;52(2):155-66. doi: 10.1002/mc.21841. Epub 2011 Nov 28. Mol Carcinog. 2013. PMID: 22121102 Free PMC article.
-
Diagnostic and prognostic value of WNT family gene expression in hepatitis B virus‑related hepatocellular carcinoma.Oncol Rep. 2019 Sep;42(3):895-910. doi: 10.3892/or.2019.7224. Epub 2019 Jul 5. Oncol Rep. 2019. PMID: 31322232 Free PMC article.
-
The JAK-STAT signaling-related signature serves as a prognostic and predictive biomarker for renal cell carcinoma immunotherapy.Gene. 2024 Nov 15;927:148719. doi: 10.1016/j.gene.2024.148719. Epub 2024 Jun 23. Gene. 2024. PMID: 38917875
-
Role of microRNAs in the main molecular pathways of hepatocellular carcinoma.World J Gastroenterol. 2018 Jul 7;24(25):2647-2660. doi: 10.3748/wjg.v24.i25.2647. World J Gastroenterol. 2018. PMID: 29991871 Free PMC article. Review.
-
Association of STAT3 and STAT4 polymorphisms with susceptibility to chronic hepatitis B virus infection and risk of hepatocellular carcinoma: a meta-analysis.Biosci Rep. 2019 Jun 20;39(6):BSR20190783. doi: 10.1042/BSR20190783. Print 2019 Jun 28. Biosci Rep. 2019. PMID: 31160486 Free PMC article.
Cited by
-
Exploring the JAK/STAT Signaling Pathway in Hepatocellular Carcinoma: Unraveling Signaling Complexity and Therapeutic Implications.Int J Mol Sci. 2023 Sep 6;24(18):13764. doi: 10.3390/ijms241813764. Int J Mol Sci. 2023. PMID: 37762066 Free PMC article. Review.
-
Identification of hepatocellular carcinoma prognostic markers based on 10-immune gene signature.Biosci Rep. 2020 Aug 28;40(8):BSR20200894. doi: 10.1042/BSR20200894. Biosci Rep. 2020. PMID: 32789471 Free PMC article.
-
JAK3 and TYK2 Serve as Prognostic Biomarkers and Are Associated with Immune Infiltration in Stomach Adenocarcinoma.Biomed Res Int. 2020 Sep 19;2020:7973568. doi: 10.1155/2020/7973568. eCollection 2020. Biomed Res Int. 2020. PMID: 33083484 Free PMC article.
-
Liver Tumor Microenvironment.Adv Exp Med Biol. 2020;1296:227-241. doi: 10.1007/978-3-030-59038-3_14. Adv Exp Med Biol. 2020. PMID: 34185296
-
Anti-Hepatocellular Carcinoma Biomolecules: Molecular Targets Insights.Int J Mol Sci. 2021 Oct 6;22(19):10774. doi: 10.3390/ijms221910774. Int J Mol Sci. 2021. PMID: 34639131 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

