Asexual reproduction reduces transposable element load in experimental yeast populations
- PMID: 31486772
- PMCID: PMC6783261
- DOI: 10.7554/eLife.48548
Asexual reproduction reduces transposable element load in experimental yeast populations
Abstract
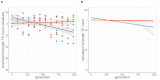
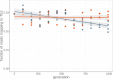
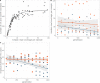
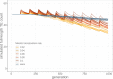
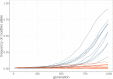
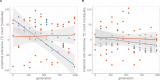
Theory predicts that sexual reproduction can either facilitate or restrain transposable element (TE) accumulation by providing TEs with a means of spreading to all individuals in a population, versus facilitating TE load reduction via purifying selection. By quantifying genomic TE loads over time in experimental sexual and asexual Saccharomyces cerevisiae populations, we provide direct evidence that TE loads decrease rapidly under asexual reproduction. We show, using simulations, that this reduction may occur via evolution of TE activity, most likely via increased excision rates. Thus, sex is a major driver of genomic TE loads and at the root of the success of TEs.
Keywords: S. cerevisiae; asexuality; evolution of sex; evolutionary biology; transposable elements.
© 2019, Bast et al.
Conflict of interest statement
JB, KJ, DS, DR, TS No competing interests declared
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
- BA 5800/1-1/Deutsche Forschungsgemeinschaft/International
- PP00P3_17062/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung/International
- PP00P3_139013/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung/International
- BA 5800/2-1/Deutsche Forschungsgemeinschaft/International
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

