Large miRNA survival analysis reveals a prognostic four-biomarker signature for triple negative breast cancer
- PMID: 31487369
- PMCID: PMC7198019
- DOI: 10.1590/1678-4685-GMB-2018-0269
Large miRNA survival analysis reveals a prognostic four-biomarker signature for triple negative breast cancer
Abstract
Triple negative breast cancer (TNBC) is currently the only major breast tumor subtype without effective targeted therapy and, as a consequence, usually presents a poor outcome. Due to its more aggressive phenotype, there is an urgent clinical need to identify novel biomarkers that discriminate individuals with poor prognosis. We hypothesize that miRNAs can be used to this end because they are involved in the initiation and progression of tumors by altering the expression of their target genes. To identify a prognostic biomarker in TNBC, we analyzed the miRNA expression of a cohort composed of 185 patients diagnosed with TNBC using penalized Cox regression models. We identified a four-biomarker signature based on miR-221, miR-1305, miR-4708, and RMDN2 expression levels that allowed for the subdivision of TNBC into high- or low-risk groups (Hazard Ratio - HR = 0.32; 95% Confidence Interval - CI = 0.11-0.91; p = 0.03) and are also statistically associated with survival outcome in subgroups of postmenopausal status (HR = 0.19; 95% CI = 0.04-0.90; p= 0.016), node negative status (HR = 0.12; 95% CI = 0.01-1.04; p = 0.026), and tumors larger than 2cm (HR = 0.21; 95% CI = 0.05-0.81; p = 0.021). This four-biomarker signature was significantly associated with TNBC as an independent prognostic factor for survival.
Figures
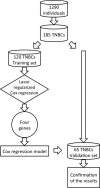
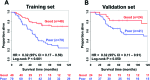

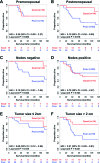
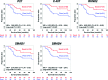

Similar articles
-
A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients.Clin Cancer Res. 2015 Mar 1;21(5):1207-14. doi: 10.1158/1078-0432.CCR-14-2011. Epub 2014 Dec 29. Clin Cancer Res. 2015. PMID: 25547678
-
miR-629-3p may serve as a novel biomarker and potential therapeutic target for lung metastases of triple-negative breast cancer.Breast Cancer Res. 2017 Jun 19;19(1):72. doi: 10.1186/s13058-017-0865-y. Breast Cancer Res. 2017. PMID: 28629464 Free PMC article.
-
CYPOR is a novel and independent prognostic biomarker of recurrence-free survival in triple-negative breast cancer patients.Int J Cancer. 2019 Feb 1;144(3):631-640. doi: 10.1002/ijc.31798. Epub 2018 Nov 7. Int J Cancer. 2019. PMID: 30110125
-
Beyond Axillary Lymph Node Metastasis, BMI and Menopausal Status Are Prognostic Determinants for Triple-Negative Breast Cancer Treated by Neoadjuvant Chemotherapy.PLoS One. 2015 Dec 18;10(12):e0144359. doi: 10.1371/journal.pone.0144359. eCollection 2015. PLoS One. 2015. PMID: 26684197 Free PMC article.
-
MicroRNAs Involved in Carcinogenesis, Prognosis, Therapeutic Resistance and Applications in Human Triple-Negative Breast Cancer.Cells. 2019 Nov 22;8(12):1492. doi: 10.3390/cells8121492. Cells. 2019. PMID: 31766744 Free PMC article. Review.
Cited by
-
The Ambivalent Role of miRNAs in Carcinogenesis: Involvement in Renal Cell Carcinoma and Their Clinical Applications.Pharmaceuticals (Basel). 2021 Apr 2;14(4):322. doi: 10.3390/ph14040322. Pharmaceuticals (Basel). 2021. PMID: 33918154 Free PMC article. Review.
-
The Role of MicroRNA as Clinical Biomarkers for Breast Cancer Surgery and Treatment.Int J Mol Sci. 2021 Aug 1;22(15):8290. doi: 10.3390/ijms22158290. Int J Mol Sci. 2021. PMID: 34361056 Free PMC article. Review.
-
Oncoinformatic screening of the gene clusters involved in the HER2-positive breast cancer formation along with the in silico pharmacodynamic profiling of selective long-chain omega-3 fatty acids as the metastatic antagonists.Mol Divers. 2023 Dec;27(6):2651-2672. doi: 10.1007/s11030-022-10573-8. Epub 2022 Nov 29. Mol Divers. 2023. PMID: 36445532
-
Effects of IncRNA PROX1-AS1 on Proliferation, Migration, Invasion and Apoptosis of Lung Cancer Cells by Regulating MiR-1305.J Healthc Eng. 2022 Mar 2;2022:9570900. doi: 10.1155/2022/9570900. eCollection 2022. J Healthc Eng. 2022. Retraction in: J Healthc Eng. 2023 Aug 16;2023:9869418. doi: 10.1155/2023/9869418. PMID: 35281529 Free PMC article. Retracted.
-
Circ0120816 acts as an oncogene of esophageal squamous cell carcinoma by inhibiting miR-1305 and releasing TXNRD1.Cancer Cell Int. 2020 Oct 29;20(1):526. doi: 10.1186/s12935-020-01617-w. Cancer Cell Int. 2020. PMID: 33292234 Free PMC article.
References
-
- Aicher A, Heeschen C, Mildner-Rihm C, Urbich C, Ihling C, Technau-Ihling K, Zeiher AM, Dimmeler S. Essential role of endothelial nitric oxide synthase for mobilization of stem and progenitor cells. Nat Med. 2003;9:1370. - PubMed
-
- Ariazi EA, Ariazi JL, Cordera F, Jordan VC. Estrogen receptors as therapeutic targets in breast cancer. Curr Top Med Chem. 2006;6:181–202. - PubMed
-
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Internet Resources
-
- European Genome-phenome Archive. [accessed 21 August 2014]. https://www.ebi.ac.uk/ega/home.
LinkOut - more resources
Full Text Sources
Other Literature Sources

