Analytical Evaluation of the Human Papillomavirus HPV DNA Array E1-Based Genotyping Assay
- PMID: 31487743
- PMCID: PMC6878751
- DOI: 10.1159/000502207
Analytical Evaluation of the Human Papillomavirus HPV DNA Array E1-Based Genotyping Assay
Abstract
Background: Cervical cancer is caused by a persistent infection of human papillomavirus (HPV). Therefore, tests which detect the carcinogenic virus can be used for cervical cancer screening.
Objective: This is the first evaluation of the HPV DNA Array (AID Diagnostika, Strassberg, Germany), an E1-based genotyping polymerase chain reaction (PCR) test for identification of 29 HPV types (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 67, 68, 69, 70, 73, 82, 85, and 97).
Methods: Analytical performance of the assay was assessed with cervical cancer cell lines with known HPV status, and preselected clinical cervical scrapings genotyped by multiplexed genotyping (MPG) with a Luminex readout (validated in-house assay). Intra- and inter-laboratory reproducibility experiments were performed to ensure the reliability of the assay.
Results: HPV DNA Array identified the intrinsic HPV genotype in all cervical cancer cell lines and demonstrated a high sensitivity for HPV16 probe (1 cell per PCR reaction), as well as HPV18 and 45 probes (100 cells per PCR reaction). When compared with MPG, HPV DNA Array showed a good agreement of 92.2% for HPV detection irrespective of type (κ = 0.601), and demonstrated high agreement for HPV16 (80.7%, κ = 0.836) and HPV18 (86.7%, κ = 0.925). Furthermore, high intra-/inter-laboratory reproducibility was observed (90.9-100%).
Conclusion: HPV DNA Array showed high sensitivity for correct HPV genotype detection in experimental and clinical samples with a good correlation to the reference test. Since HPV DNA Array is based on a simple multiplexed PCR followed by reverse hybridization in a 96-well format and automated visual readout by AID ELISpot reader, it is capable of high throughput in a time-effective manner. HPV DNA Array could be considered for extended HPV genotyping of cervical smears.
Keywords: Cervical cancer; GP5+/6+; HPV detection; Human papillomavirus; Luminex; Multiplex; Validation.
© 2019 The Author(s) Published by S. Karger AG, Basel.
Conflict of interest statement
A.P. received travel grants from AID/GeinID. M.H. and R.P. are employed at AID/GenID. AID/GenID provided the necessary kits free of charge. They had no role in study design, data collection, or analysis.
Figures
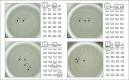
References
-
- Ho GY, Bierman R, Beardsley L, Chang CJ, Burk RD. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998 Feb;338((7)):423–8. - PubMed
-
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004 Jun;324((1)):17–27. - PubMed
-
- Garland SM, Steben M, Sings HL, James M, Lu S, Railkar R, et al. Natural history of genital warts: analysis of the placebo arm of 2 randomized phase III trials of a quadrivalent human papillomavirus (types 6, 11, 16, and 18) vaccine. J Infect Dis. 2009 Mar;199((6)):805–14. - PubMed
-
- Clifford G, Franceschi S, Diaz M, Munoz N, Villa LL. Chapter 3: HPV type-distribution in women with and without cervical neoplastic diseases. Vaccine, 2006;24(Suppl 3):p. S3/26-34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

