Molecular Dynamics Gives New Insights into the Glucose Tolerance and Inhibition Mechanisms on β-Glucosidases
- PMID: 31487855
- PMCID: PMC6766793
- DOI: 10.3390/molecules24183215
Molecular Dynamics Gives New Insights into the Glucose Tolerance and Inhibition Mechanisms on β-Glucosidases
Abstract
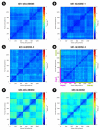
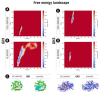
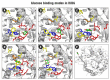
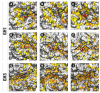
β-Glucosidases are enzymes with high importance for many industrial processes, catalyzing the last and limiting step of the conversion of lignocellulosic material into fermentable sugars for biofuel production. However, β-glucosidases are inhibited by high concentrations of the product (glucose), which limits the biofuel production on an industrial scale. For this reason, the structural mechanisms of tolerance to product inhibition have been the target of several studies. In this study, we performed in silico experiments, such as molecular dynamics (MD) simulations, free energy landscape (FEL) estimate, Poisson-Boltzmann surface area (PBSA), and grid inhomogeneous solvation theory (GIST) seeking a better understanding of the glucose tolerance and inhibition mechanisms of a representative GH1 β-glucosidase and a GH3 one. Our results suggest that the hydrophobic residues Y180, W350, and F349, as well the polar one D238 act in a mechanism for glucose releasing, herein called "slingshot mechanism", dependent also on an allosteric channel (AC). In addition, water activity modulation and the protein loop motions suggest that GH1 β-Glucosidases present an active site more adapted to glucose withdrawal than GH3, in consonance with the GH1s lower product inhibition. The results presented here provide directions on the understanding of the molecular mechanisms governing inhibition and tolerance to the product in β-glucosidases and can be useful for the rational design of optimized enzymes for industrial interests.
Keywords: GH1; GH3; Poisson–Boltzmann surface area; allosteric channel; free energy landscape; glucose tolerance; grid inhomogeneous solvation theory; molecular dynamics simulation; slingshot mechanism; β-Glucosidases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Glutantβase: a database for improving the rational design of glucose-tolerant β-glucosidases.BMC Mol Cell Biol. 2020 Jul 1;21(1):50. doi: 10.1186/s12860-020-00293-y. BMC Mol Cell Biol. 2020. PMID: 32611314 Free PMC article.
-
Thermostabilizing mechanisms of canonical single amino acid substitutions at a GH1 β-glucosidase probed by multiple MD and computational approaches.Proteins. 2023 Feb;91(2):218-236. doi: 10.1002/prot.26424. Epub 2022 Oct 10. Proteins. 2023. PMID: 36114781
-
Structural insight into the fungal β-glucosidases and their interactions with organics.Int J Biol Macromol. 2019 Oct 1;138:1019-1028. doi: 10.1016/j.ijbiomac.2019.07.177. Epub 2019 Jul 26. Int J Biol Macromol. 2019. PMID: 31356936
-
Glucose tolerant and glucose stimulated β-glucosidases - A review.Bioresour Technol. 2018 Nov;267:704-713. doi: 10.1016/j.biortech.2018.07.137. Epub 2018 Jul 27. Bioresour Technol. 2018. PMID: 30093225 Review.
-
Unconventional β-Glucosidases: A Promising Biocatalyst for Industrial Biotechnology.Appl Biochem Biotechnol. 2021 Sep;193(9):2993-3016. doi: 10.1007/s12010-021-03568-y. Epub 2021 Apr 19. Appl Biochem Biotechnol. 2021. PMID: 33871765 Review.
Cited by
-
Real-Time Measurement of Cellobiose and Glucose Formation during Enzymatic Biomass Hydrolysis.Anal Chem. 2021 Jun 1;93(21):7732-7738. doi: 10.1021/acs.analchem.1c01182. Epub 2021 May 20. Anal Chem. 2021. PMID: 34014659 Free PMC article.
-
Glutantβase: a database for improving the rational design of glucose-tolerant β-glucosidases.BMC Mol Cell Biol. 2020 Jul 1;21(1):50. doi: 10.1186/s12860-020-00293-y. BMC Mol Cell Biol. 2020. PMID: 32611314 Free PMC article.
-
VTR: A Web Tool for Identifying Analogous Contacts on Protein Structures and Their Complexes.Front Bioinform. 2021 Nov 8;1:730350. doi: 10.3389/fbinf.2021.730350. eCollection 2021. Front Bioinform. 2021. PMID: 36303745 Free PMC article.
-
Computational Insights into Glucose Tolerance and Stimulation in a Family 1 β-glucosidase.J Chem Inf Model. 2025 Jul 14;65(13):7102-7112. doi: 10.1021/acs.jcim.5c00922. Epub 2025 Jun 30. J Chem Inf Model. 2025. PMID: 40589032 Free PMC article.
-
Proteus: An algorithm for proposing stabilizing mutation pairs based on interactions observed in known protein 3D structures.BMC Bioinformatics. 2020 Jul 1;21(1):275. doi: 10.1186/s12859-020-03575-6. BMC Bioinformatics. 2020. PMID: 32611389 Free PMC article.
References
-
- Swangkeaw J., Vichitphan S., Butzke C.E., Vichitphan K. Characterization of β-glucosidases from Hanseniaspora sp. and Pichia anomala with potentially aroma-enhancing capabilities in juice and wine. World J. Microbiol. Biotechnol. 2010;27:423–430. doi: 10.1007/s11274-010-0474-8. - DOI
-
- Mariano D.C.B., Leite C., Santos L.H.S., Marins L.F., Machado K.S., Werhli A.V., Lima L.H.F., de Melo-Minardi R.C. Characterization of glucose-tolerant β-glucosidases used in biofuel production under the bioinformatics perspective: A systematic review. Genet. Mol. Res. 2017;16:1–19. doi: 10.4238/gmr16039740. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

