RNA-Seq de Novo Assembly and Differential Transcriptome Analysis of Chaga (Inonotus obliquus) Cultured with Different Betulin Sources and the Regulation of Genes Involved in Terpenoid Biosynthesis
- PMID: 31487924
- PMCID: PMC6770048
- DOI: 10.3390/ijms20184334
RNA-Seq de Novo Assembly and Differential Transcriptome Analysis of Chaga (Inonotus obliquus) Cultured with Different Betulin Sources and the Regulation of Genes Involved in Terpenoid Biosynthesis
Abstract
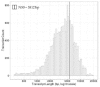
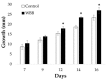
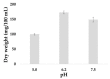
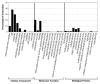
Chaga (Inonotus obliquus) is a medicinal fungus used in traditional medicine of Native American and North Eurasian cultures. Several studies have demonstrated the medicinal properties of chaga's bioactive molecules. For example, several terpenoids (e.g., betulin, betulinic acid and inotodiol) isolated from I. obliquus cells have proven effectiveness in treating different types of tumor cells. However, the molecular mechanisms and regulation underlying the biosynthesis of chaga terpenoids remain unknown. In this study, we report on the optimization of growing conditions for cultured I. obliquus in presence of different betulin sources (e.g., betulin or white birch bark). It was found that better results were obtained for a liquid culture pH 6.2 at 28 °C. In addition, a de novo assembly and characterization of I. obliquus transcriptome in these growth conditions using Illumina technology was performed. A total of 219,288,500 clean reads were generated, allowing for the identification of 20,072 transcripts of I. obliquus including transcripts involved in terpenoid biosynthesis. The differential expression of these genes was confirmed by quantitative-PCR. This study provides new insights on the molecular mechanisms and regulation of I. obliquus terpenoid production. It also contributes useful molecular resources for gene prediction or the development of biotechnologies for the alternative production of terpenoids.
Keywords: Inonotus obliquus; RNA-Seq; betulinic acid; biosynthesis; chaga; de novo transcriptome; specialized metabolism; terpenoid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Chaga ( Inonotus obliquus), a Future Potential Medicinal Fungus in Oncology? A Chemical Study and a Comparison of the Cytotoxicity Against Human Lung Adenocarcinoma Cells (A549) and Human Bronchial Epithelial Cells (BEAS-2B).Integr Cancer Ther. 2018 Sep;17(3):832-843. doi: 10.1177/1534735418757912. Epub 2018 Feb 27. Integr Cancer Ther. 2018. PMID: 29484963 Free PMC article.
-
Stimulated production of steroids in Inonotus obliquus by host factors from birch.J Biosci Bioeng. 2014 Dec;118(6):728-31. doi: 10.1016/j.jbiosc.2014.05.022. Epub 2014 Jul 12. J Biosci Bioeng. 2014. PMID: 25027706
-
Chemical Content and Cytotoxic Activity on Various Cancer Cell Lines of Chaga (Inonotus obliquus) Growing on Betula pendula and Betula pubescens.Pharmaceuticals (Basel). 2024 Aug 1;17(8):1013. doi: 10.3390/ph17081013. Pharmaceuticals (Basel). 2024. PMID: 39204121 Free PMC article.
-
A brief overview of the medicinal and nutraceutical importance of Inonotus obliquus (chaga) mushrooms.Heliyon. 2024 Aug 6;10(15):e35638. doi: 10.1016/j.heliyon.2024.e35638. eCollection 2024 Aug 15. Heliyon. 2024. PMID: 39170453 Free PMC article. Review.
-
Chemical diversity of biologically active metabolites in the sclerotia of Inonotus obliquus and submerged culture strategies for up-regulating their production.Appl Microbiol Biotechnol. 2010 Jul;87(4):1237-54. doi: 10.1007/s00253-010-2682-4. Epub 2010 Jun 8. Appl Microbiol Biotechnol. 2010. PMID: 20532760 Review.
Cited by
-
Stimulatory Effects of Oleci Acid and Fungal Elicitor on Betulinic Acid Production by Submerged Cultivation of Medicinal Mushroom Inonotus obliquus.J Fungi (Basel). 2021 Mar 31;7(4):266. doi: 10.3390/jof7040266. J Fungi (Basel). 2021. PMID: 33807450 Free PMC article.
-
Biosynthesis and regulation of terpenoids from basidiomycetes: exploration of new research.AMB Express. 2021 Nov 15;11(1):150. doi: 10.1186/s13568-021-01304-7. AMB Express. 2021. PMID: 34779947 Free PMC article. Review.
-
Cloning and functional characterization of sesquiterpene synthase genes from Inonotus obliquus using a Saccharomyces cerevisiae expression system.World J Microbiol Biotechnol. 2025 Jan 30;41(2):56. doi: 10.1007/s11274-025-04274-1. World J Microbiol Biotechnol. 2025. PMID: 39883208
-
Independent evolution of betulin biosynthesis in Inonotus obliquus.Sci Rep. 2025 Jul 1;15(1):21319. doi: 10.1038/s41598-025-05414-1. Sci Rep. 2025. PMID: 40594539 Free PMC article.
-
Comparison of Bioactive Secondary Metabolites and Cytotoxicity of Extracts from Inonotus obliquus Isolates from Different Host Species.Molecules. 2023 Jun 22;28(13):4907. doi: 10.3390/molecules28134907. Molecules. 2023. PMID: 37446570 Free PMC article.
References
-
- Glamočlija J., Ćirić A., Nikolić M., Fernandes Â., Barros L., Calhelha R.C., Ferreira I.C., Soković M., van Griensven L.J. Chemical characterization and biological activity of Chaga (Inonotus obliquus), a medicinal “mushroom”. J. Ethnopharmacol. 2015;162:323–332. doi: 10.1016/j.jep.2014.12.069. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

