Function and Transcriptional Regulation of Bovine TORC2 Gene in Adipocytes: Roles of C/EBP, XBP1, INSM1 and ZNF263
- PMID: 31487963
- PMCID: PMC6769628
- DOI: 10.3390/ijms20184338
Function and Transcriptional Regulation of Bovine TORC2 Gene in Adipocytes: Roles of C/EBP, XBP1, INSM1 and ZNF263
Abstract
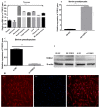
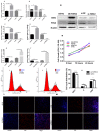
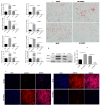
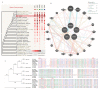
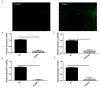
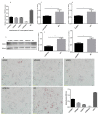
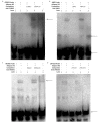
The TORC2 gene is a member of the transducer of the regulated cyclic adenosine monophosphate (cAMP) response element binding protein gene family, which plays a key role in metabolism and adipogenesis. In the present study, we confirmed the role of TORC2 in bovine preadipocyte proliferation through cell cycle staining flow cytometry, cell counting assay, 5-ethynyl-2'-deoxyuridine staining (EdU), and mRNA and protein expression analysis of proliferation-related marker genes. In addition, Oil red O staining analysis, immunofluorescence of adiponectin, mRNA and protein level expression of lipid related marker genes confirmed the role of TORC2 in the regulation of bovine adipocyte differentiation. Furthermore, the transcription start site and sub-cellular localization of the TORC2 gene was identified in bovine adipocytes. To investigate the underlying regulatory mechanism of the bovine TORC2, we cloned a 1990 bp of the 5' untranslated region (5'UTR) promoter region into a luciferase reporter vector and seven vector fragments were constructed through serial deletion of the 5'UTR flanking region. The core promoter region of the TORC2 gene was identified at location -314 to -69 bp upstream of the transcription start site. Based on the results of the transcriptional activities of the promoter vector fragments, luciferase activities of mutated fragments and siRNAs interference, four transcription factors (CCAAT/enhancer-binding protein C/BEP, X-box binding protein 1 XBP1, Insulinoma-associated 1 INSM1, and Zinc finger protein 263 ZNF263) were identified as the transcriptional regulators of TORC2 gene. These findings were further confirmed through Electrophoretic Mobility Shift Assay (EMSA) within nuclear extracts of bovine adipocytes. Furthermore, we also identified that C/EBP, XBP1, INSM1 and ZNF263 regulate TORC2 gene as activators in the promoter region. We can conclude that TORC2 gene is potentially a positive regulator of adipogenesis. These findings will not only provide an insight for the improvement of intramuscular fat in cattle, but will enhance our understanding regarding therapeutic intervention of metabolic syndrome and obesity in public health as well.
Keywords: DNA-Protein interaction; TORC2; adipogenesis; bovine adipocytes; gene regulation; intramuscular fat; luciferase reporter assay; nuclear protein; preadipocytes proliferation and differentiation; transcription factors.
Conflict of interest statement
The authors declare no conflict of interest
Figures




References
-
- Martins T.S., Sanglard L.M.P., Silva W., Chizzotti M.L., Renno L.N., Serao N.V.L., Silva F.F., Guimaraes S.E.F., Ladeira M.M., Dodson M.V., et al. Molecular Factors Underlying the Deposition of Intramuscular Fat and Collagen in Skeletal Muscle of Nellore and Angus Cattle. PLoS ONE. 2015;10:e0139943. doi: 10.1371/journal.pone.0139943. - DOI - PMC - PubMed
-
- Sasaki Y., Nagai K., Nagata Y., Doronbekov K., Nishimura S., Yoshioka S., Fujita T., Shiga K., Miyake T., Taniguchi Y., et al. Exploration of Genes Showing Intramuscular Fat Deposition-Associated Expression Changes in Musculus Longissimus Muscle. Anim. Genet. 2006;37:40–46. doi: 10.1111/j.1365-2052.2005.01380.x. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

