Molecular basis and biological function of variability in spatial genome organization
- PMID: 31488662
- PMCID: PMC7421438
- DOI: 10.1126/science.aaw9498
Molecular basis and biological function of variability in spatial genome organization
Abstract
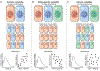
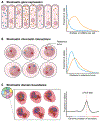
The complex three-dimensional organization of genomes in the cell nucleus arises from a wide range of architectural features including DNA loops, chromatin domains, and higher-order compartments. Although these features are universally present in most cell types and tissues, recent single-cell biochemistry and imaging approaches have demonstrated stochasticity in transcription and high variability of chromatin architecture in individual cells. We review the occurrence, mechanistic basis, and functional implications of stochasticity in genome organization. We summarize recent observations on cell- and allele-specific variability of genome architecture, discuss the nature of extrinsic and intrinsic sources of variability in genome organization, and highlight potential implications of structural heterogeneity for genome function.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

