Cushing's syndrome driver mutation disrupts protein kinase A allosteric network, altering both regulation and substrate specificity
- PMID: 31489371
- PMCID: PMC6713507
- DOI: 10.1126/sciadv.aaw9298
Cushing's syndrome driver mutation disrupts protein kinase A allosteric network, altering both regulation and substrate specificity
Abstract
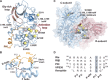
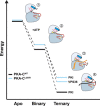
Genetic alterations in the PRKACA gene coding for the catalytic α subunit of the cAMP-dependent protein kinase A (PKA-C) are linked to cortisol-secreting adrenocortical adenomas, resulting in Cushing's syndrome. Among those, a single mutation (L205R) has been found in up to 67% of patients. Because the x-ray structures of the wild-type and mutant kinases are essentially identical, the mechanism explaining aberrant function of this mutant remains under active debate. Using NMR spectroscopy, thermodynamics, kinetic assays, and molecular dynamics simulations, we found that this single mutation causes global changes in the enzyme, disrupting the intramolecular allosteric network and eliciting losses in nucleotide/pseudo-substrate binding cooperativity. Remarkably, by rewiring its internal allosteric network, PKA-CL205R is able to bind and phosphorylate non-canonical substrates, explaining its changes in substrate specificity. Both the lack of regulation and change in substrate specificity reveal the complex role of this mutated kinase in the formation of cortisol-secreting adrenocortical adenomas.
Figures




References
-
- Lodish M., Stratakis C. A., A genetic and molecular update on adrenocortical causes of Cushing syndrome. Nat. Rev. Endocrinol. 12, 255–262 (2016). - PubMed
-
- Cao Y., He M., Gao Z., Peng Y., Li Y., Li L., Zhou W., Li X., Zhong X., Lei Y., Su T., Wang H., Jiang Y., Yang L., Wei W., Yang X., Jiang X., Liu L., He J., Ye J., Wei Q., Li Y., Wang W., Wang J., Ning G., Activating hotspot L205r mutation in PRKACA and adrenal Cushing's syndrome. Science 344, 913–917 (2014). - PubMed
-
- Sato Y., Maekawa S., Ishii R., Sanada M., Morikawa T., Shiraishi Y., Yoshida K., Nagata Y., Sato-Otsubo A., Yoshizato T., Suzuki H., Shiozawa Y., Kataoka K., Kon A., Aoki K., Chiba K., Tanaka H., Kume H., Miyano S., Fukayama M., Nureki O., Homma Y., Ogawa S., Recurrent somatic mutations underlie corticotropin-independent Cushing’s syndrome. Science 344, 917–920 (2014). - PubMed
-
- Goh G., Scholl U. I., Healy J. M., Choi M., Prasad M. L., Nelson-Williams C., Kunstman J. W., Korah R., Suttorp A.-C., Dietrich D., Haase M., Willenberg H. S., Stålberg P., Hellman P., Åkerström G., Björklund P., Carling T., Lifton R. P., Recurrent activating mutation in PRKACA in cortisol-producing adrenal tumors. Nat. Genet. 46, 613–617 (2014). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

