Metagenomic Evaluation of Bacterial and Fungal Assemblages Enriched within Diffusion Chambers and Microbial Traps Containing Uraniferous Soils
- PMID: 31489900
- PMCID: PMC6780890
- DOI: 10.3390/microorganisms7090324
Metagenomic Evaluation of Bacterial and Fungal Assemblages Enriched within Diffusion Chambers and Microbial Traps Containing Uraniferous Soils
Abstract
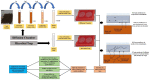
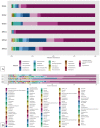
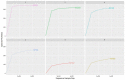
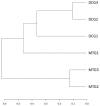
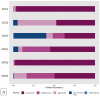
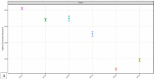
Despite significant technological advancements in the field of microbial ecology, cultivation and subsequent isolation of the vast majority of environmental microorganisms continues to pose challenges. Isolation of the environmental microbiomes is prerequisite to better understand a myriad of ecosystem services they provide, such as bioremediation of contaminants. Towards this end, in this culturomics study, we evaluated the colonization of soil bacterial and fungal communities within diffusion chambers (DC) and microbial traps (MT) established using uraniferous soils collected from a historically contaminated soil from Aiken, USA. Microbial assemblages were compared between the DC and MT relative to the native soils using amplicon based metagenomic and bioinformatic analysis. The overall rationale of this study is that DC and MT growth chambers provide the optimum conditions under which desired microbiota, identified in a previous study to serve as the "core" microbiomes, will proliferate, leading to their successful isolation. Specifically, the core microbiomes consisted of assemblages of bacteria (Burkholderia spp.) and fungi (Penicillium spp.), respectively. The findings from this study further supported previous data such that the abundance and diversity of the desired "core" microbiomes significantly increased as a function of enrichments over three consecutive generations of DC and MT, respectively. Metagenomic analysis of the DC/MT generations also revealed that enrichment and stable populations of the desired "core" bacterial and fungal microbiomes develop within the first 20 days of incubation and the practice of subsequent transfers for second and third generations, as is standard in previous studies, may be unnecessary. As a cost and time cutting measure, this study recommends running the DC/MT chambers for only a 20-day time period, as opposed to previous studies, which were run for months. In summation, it was concluded that, using the diffusion chamber-based enrichment techniques, growth of desired microbiota possessing environmentally relevant functions can be achieved in a much shorter time frame than has been previously shown.
Keywords: diffusion chamber (DC); metagenomics; microbial trap (MT); uranium.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

