Banking with precision: transfusion medicine as a potential universal application in clinical genomics
- PMID: 31490317
- PMCID: PMC7302862
- DOI: 10.1097/MOH.0000000000000536
Banking with precision: transfusion medicine as a potential universal application in clinical genomics
Abstract
Purpose of review: To summarize the most recent scientific progress in transfusion medicine genomics and discuss its role within the broad genomic precision medicine model, with a focus on the unique computational and bioinformatic aspects of this emergent field.
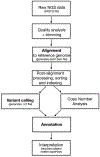
Recent findings: Recent publications continue to validate the feasibility of using next-generation sequencing (NGS) for blood group prediction with three distinct approaches: exome sequencing, whole genome sequencing, and PCR-based targeted NGS methods. The reported correlation of NGS with serologic and alternative genotyping methods ranges from 92 to 99%. NGS has demonstrated improved detection of weak antigens, structural changes, copy number variations, novel genomic variants, and microchimerism. Addition of a transfusion medicine interpretation to any clinically sequenced genome is proposed as a strategy to enhance the cost-effectiveness of precision genomic medicine. Interpretation of NGS in the blood group antigen context requires not only advanced immunohematology knowledge, but also specialized software and hardware resources, and a bioinformatics-trained workforce.
Summary: Blood transfusions are a common inpatient procedure, making blood group genomics a promising facet of precision medicine research. Further efforts are needed to embrace transfusion bioinformatic challenges and evaluate its clinical utility.
Conflict of interest statement
Conflicts of interest
There are no conflicts of interest.
Figures

Similar articles
-
Whole blood transfusion among allied partnerships: unified and interoperable blood banking for optimised care.BMJ Mil Health. 2024 Nov 25;170(6):461-464. doi: 10.1136/military-2023-002516. BMJ Mil Health. 2024. PMID: 37709507
-
Clinical Applications of Next-Generation Sequencing in Precision Oncology.Cancer J. 2019 Jul/Aug;25(4):264-271. doi: 10.1097/PPO.0000000000000385. Cancer J. 2019. PMID: 31335390 Free PMC article.
-
The impact of next-generation sequencing on genomics.J Genet Genomics. 2011 Mar 20;38(3):95-109. doi: 10.1016/j.jgg.2011.02.003. Epub 2011 Mar 15. J Genet Genomics. 2011. PMID: 21477781 Free PMC article. Review.
-
Blood banking/transfusion medicine.Clin Privil White Pap. 2013 Nov;(438):1-13. Clin Privil White Pap. 2013. PMID: 24701652 No abstract available.
-
The role of genomics in transfusion medicine.Curr Opin Hematol. 2018 Nov;25(6):509-515. doi: 10.1097/MOH.0000000000000469. Curr Opin Hematol. 2018. PMID: 30138126 Review.
Cited by
-
Hemoglobin-based transfusion strategies for cardiovascular and other diseases: restrictive, liberal, or neither?Blood. 2024 Nov 14;144(20):2075-2082. doi: 10.1182/blood.2024025927. Blood. 2024. PMID: 39293024
-
Enabling Technologies for Personalized and Precision Medicine.Trends Biotechnol. 2020 May;38(5):497-518. doi: 10.1016/j.tibtech.2019.12.021. Epub 2020 Jan 21. Trends Biotechnol. 2020. PMID: 31980301 Free PMC article. Review.
-
Application of Different Ventilation Modes Combined with AutoFlow Technology in Thoracic Surgery.J Healthc Eng. 2022 Mar 28;2022:2507149. doi: 10.1155/2022/2507149. eCollection 2022. J Healthc Eng. 2022. Retraction in: J Healthc Eng. 2023 Aug 2;2023:9812584. doi: 10.1155/2023/9812584. PMID: 35388321 Free PMC article. Retracted. Clinical Trial.
References
-
- Landsteiner K Zur Kenntnis der antifermentativen, lytischen und agglutinier- enden Wirkungen des Blutserums und der Lymphe. Zentralblatt fur Bakteriologie. 1900;27:357–62.
-
- Westhoff CM. Blood group genotyping. Blood. 2019;133(17):1814–20. - PubMed
- This is a thorough recent review on the application of blood group genotyping in various clinical contexts, including a brief discussion on NGS in transfusion.
-
- Elkins MB, Davenport RD, Bluth MH. Molecular Pathology in Transfusion Medicine: New Concepts and Applications. Clin Lab Med. 2018;38(2):277–92. - PubMed
-
- Hyland CA, Roulis EV, Schoeman EM. Developments beyond blood group serology in the genomics era. Br J Haematol. 2019;184(6):897–911. - PubMed
- This manuscript reviews the genetic basis of blood groups and the published research on transfusion medicine genomics. The article provides blood group gene coordinates in the most recent human reference build GRCh38, and it discusses future applications of NGS in the blood bank.
-
- Wheeler MM, Johnsen JM. The role of genomics in transfusion medicine. Curr Opin Hematol. 2018;25(6):509–15. - PubMed
- This article is a preceding review on transfusion medicine genomics, on which the present manuscript is built.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

