Origins of the arctic fox variant rabies viruses responsible for recent cases of the disease in southern Ontario
- PMID: 31490919
- PMCID: PMC6750613
- DOI: 10.1371/journal.pntd.0007699
Origins of the arctic fox variant rabies viruses responsible for recent cases of the disease in southern Ontario
Abstract
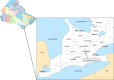
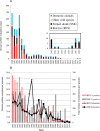
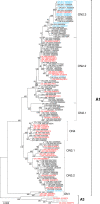
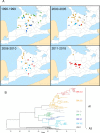
A subpopulation of the arctic fox lineage of rabies virus has circulated extensively in red fox populations of Ontario, Canada, between the 1960s and 1990s. An intensive wildlife rabies control program, in which field operations were initiated in 1989, resulted in elimination of the disease in eastern Ontario. However in southwestern Ontario, as numbers of rabid foxes declined the proportion of skunks confirmed to be infected with this rabies virus variant increased and concerted control efforts targeting this species were employed to eliminate the disease. Since 2012 no cases due to this viral variant were reported in southwestern Ontario until 2015 when a single case of rabies due to the arctic fox variant was reported in a bovine. Several additional cases have been documented subsequently. Since routine antigenic typing cannot discriminate between the variants which previously circulated in Ontario and those from northern Canada it was unknown whether these recent cases were the result of a new introduction of this variant or a continuation of the previous enzootic. To explore the origins of this new outbreak whole genome sequences of a collection of 128 rabies viruses recovered from Ontario between the 1990s to the present were compared with those representative of variants circulating in the Canadian north. Phylogenetic analysis shows that the variant responsible for current cases in southwestern Ontario has evolved from those variants known to circulate in Ontario previously and is not due to a new introduction from northern regions. Thus despite ongoing passive surveillance the persistence of wildlife rabies went undetected in the study area for almost three years. The apparent adaptation of this rabies virus variant to the skunk host provided the opportunity to explore coding changes in the viral genome which might be associated with this host shift. Several such changes were identified including a subset for which the operation of positive selection was supported. The location of a small number of these amino acid substitutions in or close to protein motifs of functional importance suggests that some of them may have played a role in this host shift.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following potential competing interests: Both authors have previously received funding from the OMNRF for studies related to wildlife rabies and its control. This association had no impact on the data collected or its interpretation as presented in this report.
Figures
References
-
- Jackson AC, Fu ZF (2013) Pathogenesis In: Jackson AC, editor. Rabies: Scientific basis of the disease and its management. Third ed. Oxford, UK: Elsevier; pp. 299–349.
-
- Wunner WH, Conzelmann K-K (2013) Rabies virus In: Jackson AC, editor. Rabies third ed. Oxford, UK: Academic Press; pp. 17–60.
-
- Steinhauer DA, Holland JJ (1987) Rapid Evolution of RNA viruses. Annual Review of Microbiology 41: 409–433. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

